Temporal and Epigenetic Control of Plasticity and Fate Decision during CD8+ T-Cell Memory Differentiation
- PMID: 33972365
- PMCID: PMC8635004
- DOI: 10.1101/cshperspect.a037754
Temporal and Epigenetic Control of Plasticity and Fate Decision during CD8+ T-Cell Memory Differentiation
Abstract
Immunological memory is a fundamental hallmark of the adaptive immune responses and one of the most relevant aspects of protective immunity. Our understanding of the processes of memory T-cell differentiation and maintenance of long-term immunity is continuously evolving, and recent advances highlight new regulatory networks and chromatin dynamic changes contributing to maintain T-cell identity and impeding the reprogramming of specific T-cell states. Here, the current understanding of the mechanisms that generate the diversity and the heterogeneity of CD8+ T-cell subsets will be discussed, focusing on the temporal and epigenetic mechanisms orchestrating the establishment and maintenance of distinct states of T-cell fate determination and functional commitment.
Copyright © 2021 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
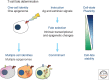
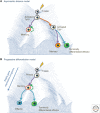

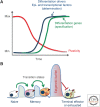
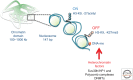
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
