Cell-free expression of the outer membrane protein OprF of Pseudomonas aeruginosa for vaccine purposes
- PMID: 33972378
- PMCID: PMC8127326
- DOI: 10.26508/lsa.202000958
Cell-free expression of the outer membrane protein OprF of Pseudomonas aeruginosa for vaccine purposes
Abstract
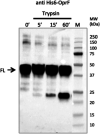
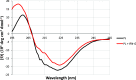
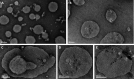
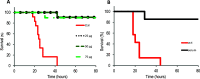
Pseudomonas aeruginosa is the second-leading cause of nosocomial infections and pneumonia in hospitals. Because of its extraordinary capacity for developing resistance to antibiotics, treating infections by Pseudomonas is becoming a challenge, lengthening hospital stays, and increasing medical costs and mortality. The outer membrane protein OprF is a well-conserved and immunogenic porin playing an important role in quorum sensing and in biofilm formation. Here, we used a bacterial cell-free expression system to reconstitute OprF under its native forms in liposomes and we demonstrated that the resulting OprF proteoliposomes can be used as a fully functional recombinant vaccine against P. aeruginosa Remarkably, we showed that our system promotes the folding of OprF into its active open oligomerized state as well as the formation of mega-pores. Our approach thus represents an easy and efficient way for producing bacterial membrane antigens exposing native epitopes for vaccine purposes.
© 2021 Mayeux et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





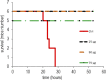
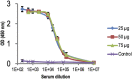
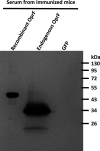

References
-
- Adlbrecht C, Wurm R, Depuydt P, Spapen H, Lorente JA, Staudinger T, Creteur J, Zauner C, Meier-Hellmann A, Eller P, et al. (2020) Efficacy, immunogenicity, and safety of IC43 recombinant Pseudomonas aeruginosa vaccine in mechanically ventilated intensive care patients-a randomized clinical trial. Crit Care 24: 74. 10.1186/s13054-020-2792-z - DOI - PMC - PubMed
-
- Bahey-El-Din M, Mohamed SA, Sheweita SA, Haroun M, Zaghloul TI (2020) Recombinant N-terminal outer membrane porin (OprF) of Pseudomonas aeruginosa is a promising vaccine candidate against both P. aeruginosa and some strains of Acinetobacter baumannii. Int J Med Microbiol 310: 151415. 10.1016/j.ijmm.2020.151415 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
