Vps34 and TOR Kinases Coordinate HAC1 mRNA Translation in the Presence or Absence of Ire1-Dependent Splicing
- PMID: 33972394
- PMCID: PMC8224233
- DOI: 10.1128/MCB.00662-20
Vps34 and TOR Kinases Coordinate HAC1 mRNA Translation in the Presence or Absence of Ire1-Dependent Splicing
Abstract
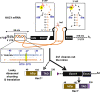
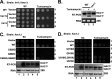
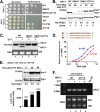
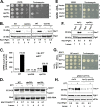
In the budding yeast Saccharomyces cerevisiae, an mRNA, called HAC1, exists in a translationally repressed form in the cytoplasm. Under conditions of cellular stress, such as when unfolded proteins accumulate inside the endoplasmic reticulum (ER), an RNase Ire1 removes an intervening sequence (intron) from the HAC1 mRNA by nonconventional cytosolic splicing. Removal of the intron results in translational derepression of HAC1 mRNA and production of a transcription factor that activates expression of many enzymes and chaperones to increase the protein-folding capacity of the cell. Here, we show that Ire1-mediated RNA cleavage requires Watson-Crick base pairs in two RNA hairpins, which are located at the HAC1 mRNA exon-intron junctions. Then, we show that the translational derepression of HAC1 mRNA can occur independent of cytosolic splicing. These results are obtained from HAC1 variants that translated an active Hac1 protein from the unspliced mRNA. Additionally, we show that the phosphatidylinositol-3-kinase Vps34 and the nutrient-sensing kinases TOR and GCN2 are key regulators of HAC1 mRNA translation and consequently the ER stress responses. Collectively, our data suggest that the cytosolic splicing and the translational derepression of HAC1 mRNA are coordinated by unique and parallel networks of signaling pathways.
Keywords: ER; Gcn2; Hac1; Ire1; TOR; UPR; Vps34; translation.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
