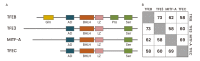
MiT Family Transcriptional Factors in Immune Cell Functions
- PMID: 33972476
- PMCID: PMC8175148
- DOI: 10.14348/molcells.2021.0067
MiT Family Transcriptional Factors in Immune Cell Functions
Abstract
The microphthalmia-associated transcription factor family (MiT family) proteins are evolutionarily conserved transcription factors that perform many essential biological functions. In mammals, the MiT family consists of MITF (microphthalmia-associated transcription factor or melanocyte-inducing transcription factor), TFEB (transcription factor EB), TFE3 (transcription factor E3), and TFEC (transcription factor EC). These transcriptional factors belong to the basic helix-loop-helix-leucine zipper (bHLH-LZ) transcription factor family and bind the E-box DNA motifs in the promoter regions of target genes to enhance transcription. The best studied functions of MiT proteins include lysosome biogenesis and autophagy induction. In addition, they modulate cellular metabolism, mitochondria dynamics, and various stress responses. The control of nuclear localization via phosphorylation and dephosphorylation serves as the primary regulatory mechanism for MiT family proteins, and several kinases and phosphatases have been identified to directly determine the transcriptional activities of MiT proteins. In different immune cell types, each MiT family member is shown to play distinct or redundant roles and we expect that there is far more to learn about their functions and regulatory mechanisms in host defense and inflammatory responses.
Keywords: MiT family transcription factors; autophagy; immune cells; lysosome; metabolism; microphthalmia-associated transcription factor (MITF); mitochondria; stress response; transcription factor E3 (TFE3); transcription factor EB (TFEB); transcription factor EC.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures

References
-
- Argani P., Reuter V.E., Zhang L., Sung Y.S., Ning Y., Epstein J.I., Netto G.J., Antonescu C.R. TFEB-amplified renal cell carcinomas: an aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am. J. Surg. Pathol. 2016;40:1484–1495. doi: 10.1097/PAS.0000000000000720. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

