Predictive performance of international COVID-19 mortality forecasting models
- PMID: 33972512
- PMCID: PMC8110547
- DOI: 10.1038/s41467-021-22457-w
Predictive performance of international COVID-19 mortality forecasting models
Abstract
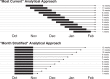
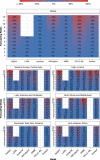
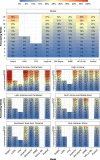
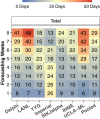
Forecasts and alternative scenarios of COVID-19 mortality have been critical inputs for pandemic response efforts, and decision-makers need information about predictive performance. We screen n = 386 public COVID-19 forecasting models, identifying n = 7 that are global in scope and provide public, date-versioned forecasts. We examine their predictive performance for mortality by weeks of extrapolation, world region, and estimation month. We additionally assess prediction of the timing of peak daily mortality. Globally, models released in October show a median absolute percent error (MAPE) of 7 to 13% at six weeks, reflecting surprisingly good performance despite the complexities of modelling human behavioural responses and government interventions. Median absolute error for peak timing increased from 8 days at one week of forecasting to 29 days at eight weeks and is similar for first and subsequent peaks. The framework and public codebase ( https://github.com/pyliu47/covidcompare ) can be used to compare predictions and evaluate predictive performance going forward.
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Predictive performance of international COVID-19 mortality forecasting models.medRxiv [Preprint]. 2020 Nov 19:2020.07.13.20151233. doi: 10.1101/2020.07.13.20151233. medRxiv. 2020. Update in: Nat Commun. 2021 May 10;12(1):2609. doi: 10.1038/s41467-021-22457-w. PMID: 33236023 Free PMC article. Updated. Preprint.
References
-
- Team, I. C.-19 health service utilization forecasting & Murray, C. J. Forecasting COVID-19 impact on hospital bed-days, ICU-days, ventilator-days and deaths by US state in the next 4 months. Preprint at medRxiv10.1101/2020.03.27.20043752 (2020).
-
- Lu, F. S., Nguyen, A. T., Link, N. B., Lipsitch, M. & Santillana, M. Estimating the early outbreak cumulative incidence of COVID-19 in the United States: three complementary approaches. Preprint at medRxiv10.1101/2020.04.18.20070821 (2020).
-
- Difan, Z. et al. Epidemic model guided machine learning for COVID-19 forecasts in the United States. Preprint at medRxivhttps://www.medrxiv.org/content/10.1101/2020.05.24.20111989v1 (2020). - DOI
-
- Ranney, M. L., Griffeth, V. & Jha, A. K. Critical supply shortages—the need for ventilators and personal protective equipment during the Covid-19 pandemic. N. Engl. J. Med. 382, e41 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

