LRIG1 is a gatekeeper to exit from quiescence in adult neural stem cells
- PMID: 33972529
- PMCID: PMC8110534
- DOI: 10.1038/s41467-021-22813-w
LRIG1 is a gatekeeper to exit from quiescence in adult neural stem cells
Abstract
Adult neural stem cells (NSCs) must tightly regulate quiescence and proliferation. Single-cell analysis has suggested a continuum of cell states as NSCs exit quiescence. Here we capture and characterize in vitro primed quiescent NSCs and identify LRIG1 as an important regulator. We show that BMP-4 signaling induces a dormant non-cycling quiescent state (d-qNSCs), whereas combined BMP-4/FGF-2 signaling induces a distinct primed quiescent state poised for cell cycle re-entry. Primed quiescent NSCs (p-qNSCs) are defined by high levels of LRIG1 and CD9, as well as an interferon response signature, and can efficiently engraft into the adult subventricular zone (SVZ) niche. Genetic disruption of Lrig1 in vivo within the SVZ NSCs leads an enhanced proliferation. Mechanistically, LRIG1 primes quiescent NSCs for cell cycle re-entry and EGFR responsiveness by enabling EGFR protein levels to increase but limiting signaling activation. LRIG1 is therefore an important functional regulator of NSC exit from quiescence.
Conflict of interest statement
S.M.P. is a founder and shareholder of Cellinta Ltd., a biotechnology startup that is developing cancer therapeutics, including glioblastoma. The other authors declare no competing interests.
Figures
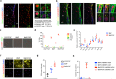
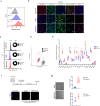
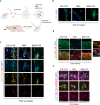

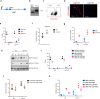

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

