Leukemia stemness and co-occurring mutations drive resistance to IDH inhibitors in acute myeloid leukemia
- PMID: 33972549
- PMCID: PMC8110775
- DOI: 10.1038/s41467-021-22874-x
Leukemia stemness and co-occurring mutations drive resistance to IDH inhibitors in acute myeloid leukemia
Abstract
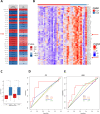
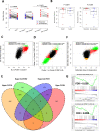
Allosteric inhibitors of mutant IDH1 or IDH2 induce terminal differentiation of the mutant leukemic blasts and provide durable clinical responses in approximately 40% of acute myeloid leukemia (AML) patients with the mutations. However, primary resistance and acquired resistance to the drugs are major clinical issues. To understand the molecular underpinnings of clinical resistance to IDH inhibitors (IDHi), we perform multipronged genomic analyses (DNA sequencing, RNA sequencing and cytosine methylation profiling) in longitudinally collected specimens from 60 IDH1- or IDH2-mutant AML patients treated with the inhibitors. The analysis reveals that leukemia stemness is a major driver of primary resistance to IDHi, whereas selection of mutations in RUNX1/CEBPA or RAS-RTK pathway genes is the main driver of acquired resistance to IDHi, along with BCOR, homologous IDH gene, and TET2. These data suggest that targeting stemness and certain high-risk co-occurring mutations may overcome resistance to IDHi in AML.
Conflict of interest statement
K.T. receives advisory and consultancy fee from Celgene, Novartis, GSK, Symbio Pharmaceuticals, and Kyowa Hakko Kirin. K.M.B. and M.F. were Celgene employee. B.W. and G.L. were Agios employee. C.D.D. receives research support (to institution) from Abbvie, Agios, Calithera, Cleave, BMS/Celgene, Daiichi-Sankyo, Forma, Loxo, and ImmuneOnc, and is among the Consultant/Advisory Boards at Abbvie, Agios, Celgene/BMS, ImmuneOnc, Novartis, Takeda, and Aprea. C.D.D. is a scientific advisor with stock options from Notable Labs. K.N.B. is a consultant for Iterion Therapeutics. E.J. receives research support (to institution) and consultancy fee from AbbVie, Amgen, Adaptive biotechnologies, Ascentage, BMS/Celgene, Genentech, Pfizer, and Takeda. H.K. receives research grants from AbbVie, Amgen, Ascentage, BMS, Daiichi-Sankyo, Immunogen, Jazz, Novartis, Pfizer and Sanofi, and honoraria from AbbVie, Actinium (Advisory Board), Adaptive Biotechnologies, Amgen, Apptitude Health, BioAscend, Daiichi-Sankyo, Delta Fly, Janssen Global, Novartis, Oxford Biometical, Pfizer and Takeda. F.R. is member of advisory boards for Celgene, BMS, and Agios, and receives honoraria from them. K.P. receives consultancy fee from Novartis. T.K. receives consulting fee from AbbVie, Agios, Daiichi-Sankyo, Genentech, Jazz Pharmaceuticals, Liberum, Novartis, Pfizer, Sanofi-Aventis, and receives research support (to institution) from AbbVie, Amgen, BMS, Genentech, Jazz Pharmaceuticals, Pfizer, Cellenkos, Ascentage, Genfleet, Astellas, and AstraZeneca, and honoraria from Genzyme. All other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

