ChAdOx1 nCoV-19 (AZD1222) vaccine candidate significantly reduces SARS-CoV-2 shedding in ferrets
- PMID: 33972565
- PMCID: PMC8110954
- DOI: 10.1038/s41541-021-00315-6
ChAdOx1 nCoV-19 (AZD1222) vaccine candidate significantly reduces SARS-CoV-2 shedding in ferrets
Abstract
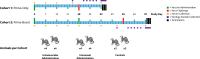
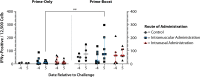
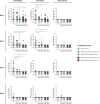
Vaccines against SARS-CoV-2 are likely to be critical in the management of the ongoing pandemic. A number of candidates are in Phase III human clinical trials, including ChAdOx1 nCoV-19 (AZD1222), a replication-deficient chimpanzee adenovirus-vectored vaccine candidate. In preclinical trials, the efficacy of ChAdOx1 nCoV-19 against SARS-CoV-2 challenge was evaluated in a ferret model of infection. Groups of ferrets received either prime-only or prime-boost administration of ChAdOx1 nCoV-19 via the intramuscular or intranasal route. All ChAdOx1 nCoV-19 administration combinations resulted in significant reductions in viral loads in nasal-wash and oral swab samples. No vaccine-associated adverse events were observed associated with the ChAdOx1 nCoV-19 candidate, with the data from this study suggesting it could be an effective and safe vaccine against COVID-19. Our study also indicates the potential for intranasal administration as a way to further improve the efficacy of this leading vaccine candidate.
Conflict of interest statement
Oxford University has entered into a partnership with AstraZeneca for further development of ChAdOx1 nCoV-19. S.C.G. is a co-founder of Vaccitech (collaborators in the early development of this vaccine candidate) and named as an inventor on a patent covering the use of ChAdOx1-vectored vaccines and a patent application covering this SARS-CoV-2 vaccine (PCT/GB2012/000467). T.L. is named as an inventor on a patent application covering this SARS-CoV-2 vaccine and was a consultant to Vaccitech for an unrelated project, during the conduct of the study. The remaining authors declare no competing interests.
Figures


References
-
- CSSE. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University.https://coronavirus.jhu.edu/map.html (2020).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

