Application of the targeted sequencing approach reveals the single nucleotide polymorphism (SNP) repertoire in microRNA genes in the pig genome
- PMID: 33972633
- PMCID: PMC8110958
- DOI: 10.1038/s41598-021-89363-5
Application of the targeted sequencing approach reveals the single nucleotide polymorphism (SNP) repertoire in microRNA genes in the pig genome
Abstract
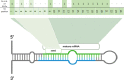
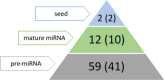
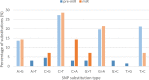
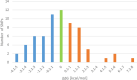
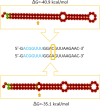
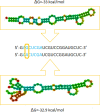
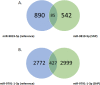
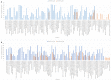
MicroRNAs (miRNAs) are recognized as gene expression regulators, indirectly orchestrating a plethora of biological processes. Single nucleotide polymorphism (SNP), one of the most common genetic variations in the genome, is established to affect miRNA functioning and influence complex traits and diseases. SNPs in miRNAs have also been associated with important production traits in livestock. Thus, the aim of our study was to reveal the SNP variability of miRNA genes in the genome of the pig, which is a significant farm animal and large-mammal human model. To this end, we applied the targeted sequencing approach, enabling deep sequencing of specified genomic regions. As a result, 73 SNPs localized in 50 distinct pre-miRNAs were identified. In silico analysis revealed that many of the identified SNPs influenced the structure and energy of the hairpin precursors. Moreover, SNPs localized in the seed regions were shown to alter targeted genes and, as a result, enrich different biological pathways. The obtained results corroborate a significant impact of SNPs on the miRNA processing and broaden the state of knowledge in the field of animal genomics. We also report the targeted sequencing approach to be a promising alternative for the whole genome sequencing in miRNA genes focused studies.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

