Endoplasmic reticulum stress regulates the intestinal stem cell state through CtBP2
- PMID: 33972635
- PMCID: PMC8111031
- DOI: 10.1038/s41598-021-89326-w
Endoplasmic reticulum stress regulates the intestinal stem cell state through CtBP2
Abstract
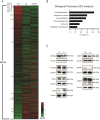
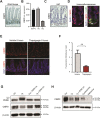
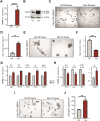
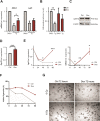
Enforcing differentiation of cancer stem cells is considered as a potential strategy to sensitize colorectal cancer cells to irradiation and chemotherapy. Activation of the unfolded protein response, due to endoplasmic reticulum (ER) stress, causes rapid stem cell differentiation in normal intestinal and colon cancer cells. We previously found that stem cell differentiation was mediated by a Protein kinase R-like ER kinase (PERK) dependent arrest of mRNA translation, resulting in rapid protein depletion of WNT-dependent transcription factor c-MYC. We hypothesize that ER stress dependent stem cell differentiation may rely on the depletion of additional transcriptional regulators with a short protein half-life that are rapidly depleted due to a PERK-dependent translational pause. Using a novel screening method, we identify novel transcription factors that regulate the intestinal stem cell fate upon ER stress. ER stress was induced in LS174T cells with thapsigargin or subtilase cytotoxin (SubAB) and immediate alterations in nuclear transcription factor activity were assessed by the CatTFRE assay in which transcription factors present in nuclear lysate are bound to plasmid DNA, co-extracted and quantified using mass-spectrometry. The role of altered activity of transcription factor CtBP2 was further examined by modification of its expression levels using CAG-rtTA3-CtBP2 overexpression in small intestinal organoids, shCtBP2 knockdown in LS174T cells, and familial adenomatous polyposis patient-derived organoids. CtBP2 overexpression organoids were challenged by ER stress and ionizing irradiation. We identified a unique set of transcription factors with altered activation upon ER stress. Gene ontology analysis showed that transcription factors with diminished binding were involved in cellular differentiation processes. ER stress decreased CtBP2 protein expression in mouse small intestine. ER stress induced loss of CtBP2 expression which was rescued by inhibition of PERK signaling. CtBP2 was overexpressed in mouse and human colorectal adenomas. Inducible CtBP2 overexpression in organoids conferred higher clonogenic potential, resilience to irradiation-induced damage and a partial rescue of ER stress-induced loss of stemness. Using an unbiased proteomics approach, we identified a unique set of transcription factors for which DNA-binding activity is lost directly upon ER stress. We continued investigating the function of co-regulator CtBP2, and show that CtBP2 mediates ER stress-induced loss of stemness which supports the intestinal stem cell state in homeostatic stem cells and colorectal cancer cells.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response.Cell Rep. 2013 Apr 25;3(4):1128-39. doi: 10.1016/j.celrep.2013.02.031. Epub 2013 Mar 28. Cell Rep. 2013. PMID: 23545496
-
PERK signaling through C/EBPδ contributes to ER stress-induced expression of immunomodulatory and tumor promoting chemokines by cancer cells.Cell Death Dis. 2021 Nov 1;12(11):1038. doi: 10.1038/s41419-021-04318-y. Cell Death Dis. 2021. PMID: 34725321 Free PMC article.
-
The kinase PERK represses translation of the G-protein-coupled receptor LGR5 and receptor tyrosine kinase ERBB3 during ER stress in cancer cells.J Biol Chem. 2020 Apr 3;295(14):4591-4603. doi: 10.1074/jbc.RA119.010655. Epub 2020 Feb 27. J Biol Chem. 2020. PMID: 32107308 Free PMC article.
-
Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate.Cell Death Dis. 2015 Jul 30;6(7):e1822. doi: 10.1038/cddis.2015.183. Cell Death Dis. 2015. PMID: 26225772 Free PMC article. Review.
-
The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress.Curr Mol Med. 2016;16(6):533-44. doi: 10.2174/1566524016666160523143937. Curr Mol Med. 2016. PMID: 27211800 Free PMC article. Review.
Cited by
-
The Implications of Cannabinoid-Induced Metabolic Dysregulation for Cellular Differentiation and Growth.Int J Mol Sci. 2023 Jul 2;24(13):11003. doi: 10.3390/ijms241311003. Int J Mol Sci. 2023. PMID: 37446181 Free PMC article. Review.
-
Alterations in ether phospholipids metabolism activate the conserved UPR-Xbp1- PDIA3/ERp60 signaling to maintain intestinal homeostasis.iScience. 2025 Feb 4;28(3):111946. doi: 10.1016/j.isci.2025.111946. eCollection 2025 Mar 21. iScience. 2025. PMID: 40034858 Free PMC article.
-
Intestinal Apc-inactivation induces HSP25 dependency.EMBO Mol Med. 2022 Dec 7;14(12):e16194. doi: 10.15252/emmm.202216194. Epub 2022 Nov 2. EMBO Mol Med. 2022. PMID: 36321561 Free PMC article.
-
C-Terminal Binding Protein 2 Emerges as a Critical Player Linking Metabolic Imbalance to the Pathogenesis of Obesity.J Atheroscler Thromb. 2024 Feb 1;31(2):109-116. doi: 10.5551/jat.RV22014. Epub 2023 Oct 3. J Atheroscler Thromb. 2024. PMID: 37793810 Free PMC article.
-
Nitrogen-doped carbon quantum dot regulates cell proliferation and differentiation by endoplasmic reticulum stress.Anim Cells Syst (Seoul). 2024 Oct 2;28(1):481-494. doi: 10.1080/19768354.2024.2409452. eCollection 2024. Anim Cells Syst (Seoul). 2024. PMID: 39364144 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

