Viruses infecting a warm water picoeukaryote shed light on spatial co-occurrence dynamics of marine viruses and their hosts
- PMID: 33972727
- PMCID: PMC8528832
- DOI: 10.1038/s41396-021-00989-9
Viruses infecting a warm water picoeukaryote shed light on spatial co-occurrence dynamics of marine viruses and their hosts
Abstract
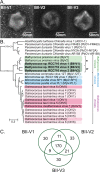
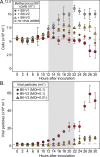
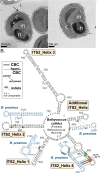
The marine picoeukaryote Bathycoccus prasinos has been considered a cosmopolitan alga, although recent studies indicate two ecotypes exist, Clade BI (B. prasinos) and Clade BII. Viruses that infect Bathycoccus Clade BI are known (BpVs), but not that infect BII. We isolated three dsDNA prasinoviruses from the Sargasso Sea against Clade BII isolate RCC716. The BII-Vs do not infect BI, and two (BII-V2 and BII-V3) have larger genomes (~210 kb) than BI-Viruses and BII-V1. BII-Vs share ~90% of their proteins, and between 65% to 83% of their proteins with sequenced BpVs. Phylogenomic reconstructions and PolB analyses establish close-relatedness of BII-V2 and BII-V3, yet BII-V2 has 10-fold higher infectivity and induces greater mortality on host isolate RCC716. BII-V1 is more distant, has a shorter latent period, and infects both available BII isolates, RCC716 and RCC715, while BII-V2 and BII-V3 do not exhibit productive infection of the latter in our experiments. Global metagenome analyses show Clade BI and BII algal relative abundances correlate positively with their respective viruses. The distributions delineate BI/BpVs as occupying lower temperature mesotrophic and coastal systems, whereas BII/BII-Vs occupy warmer temperature, higher salinity ecosystems. Accordingly, with molecular diagnostic support, we name Clade BII Bathycoccus calidus sp. nov. and propose that molecular diversity within this new species likely connects to the differentiated host-virus dynamics observed in our time course experiments. Overall, the tightly linked biogeography of Bathycoccus host and virus clades observed herein supports species-level host specificity, with strain-level variations in infection parameters.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

