Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic
- PMID: 33972780
- PMCID: PMC9248294
- DOI: 10.1038/s41588-021-00862-7
Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic
Abstract
As the SARS-CoV-2 virus spreads through human populations, the unprecedented accumulation of viral genome sequences is ushering in a new era of 'genomic contact tracing'-that is, using viral genomes to trace local transmission dynamics. However, because the viral phylogeny is already so large-and will undoubtedly grow many fold-placing new sequences onto the tree has emerged as a barrier to real-time genomic contact tracing. Here, we resolve this challenge by building an efficient tree-based data structure encoding the inferred evolutionary history of the virus. We demonstrate that our approach greatly improves the speed of phylogenetic placement of new samples and data visualization, making it possible to complete the placements under the constraints of real-time contact tracing. Thus, our method addresses an important need for maintaining a fully updated reference phylogeny. We make these tools available to the research community through the University of California Santa Cruz SARS-CoV-2 Genome Browser to enable rapid cross-referencing of information in new virus sequences with an ever-expanding array of molecular and structural biology data. The methods described here will empower research and genomic contact tracing for SARS-CoV-2 specifically for laboratories worldwide.
Figures








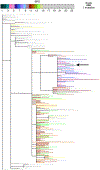

Update of
-
Ultrafast Sample Placement on Existing Trees (UShER) Empowers Real-Time Phylogenetics for the SARS-CoV-2 Pandemic.bioRxiv [Preprint]. 2020 Sep 28:2020.09.26.314971. doi: 10.1101/2020.09.26.314971. bioRxiv. 2020. Update in: Nat Genet. 2021 Jun;53(6):809-816. doi: 10.1038/s41588-021-00862-7. PMID: 33024970 Free PMC article. Updated. Preprint.
References
-
- Lam TT-Y et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
- U41 HG002371/HG/NHGRI NIH HHS/United States
- R35 GM128932/GM/NIGMS NIH HHS/United States
- 5U41HG002371-19/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)
- DP200103151/Department of Education and Training | Australian Research Council (ARC)
- T32 HG008345/HG/NHGRI NIH HHS/United States
- 2020-0000000020/Center for Information Technology Research in the Interest of Society (CITRIS)
- R35GM128932/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- T32HG008345/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)
- R01 HG010485/HG/NHGRI NIH HHS/United States
- F31HG010584/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)
- F31 HG010584/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

