This is a preprint.
ORAI1 establishes resistance to SARS-CoV-2 infection by regulating tonic type I interferon signaling
- PMID: 33972946
- PMCID: PMC8109209
- DOI: 10.1101/2021.05.04.442548
ORAI1 establishes resistance to SARS-CoV-2 infection by regulating tonic type I interferon signaling
Update in
-
ORAI1 Limits SARS-CoV-2 Infection by Regulating Tonic Type I IFN Signaling.J Immunol. 2022 Jan 1;208(1):74-84. doi: 10.4049/jimmunol.2100742. Epub 2021 Nov 24. J Immunol. 2022. PMID: 34819389 Free PMC article.
Abstract
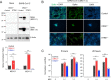
ORAI1 and STIM1 are the critical mediators of store-operated Ca 2+ entry by acting as the pore subunit and an endoplasmic reticulum-resident signaling molecule, respectively. In addition to Ca 2+ signaling, STIM1 is also involved in regulation of a cytosolic nucleic acid sensing pathway. Using ORAI1 and STIM1 knockout cells, we examined their contribution to the host response to SARS-CoV-2 infection. STIM1 knockout cells showed strong resistance to SARS-CoV-2 infection due to enhanced type I interferon response. On the contrary, ORAI1 knockout cells showed high susceptibility to SARS-CoV-2 infection as judged by increased expression of viral proteins and a high viral load. Mechanistically, ORAI1 knockout cells showed reduced homeostatic cytoplasmic Ca 2+ concentration and severe impairment in tonic interferon signaling. Transcriptome analysis showed downregulation of multiple cellular defense mechanisms, including antiviral signaling pathways in ORAI1 knockout cells, which are likely due to reduced expression of the Ca 2+ -dependent transcription factors of the activator protein 1 (AP-1) family and MEF2C . Our results identify a novel role of ORAI1-mediated Ca 2+ signaling in regulating the baseline type I interferon level, which is a determinant of host resistance to SARS-CoV-2 infection.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors do not have any competing financial interests.
Figures






References
-
- Ban N., Yamada Y., Someya Y., Ihara Y., Adachi T., Kubota A., Watanabe R., Kuroe A., Inada A., Miyawaki K., et al. (2000). Activating transcription factor-2 is a positive regulator in CaM kinase IV-induced human insulin gene expression. Diabetes 49, 1142–1148. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
