Genome-wide analysis of microRNA156 and its targets, the genes encoding SQUAMOSA promoter-binding protein-like (SPL) transcription factors, in the grass family Poaceae
- PMID: 33973419
- PMCID: PMC8110463
- DOI: 10.1631/jzus.B2000519
Genome-wide analysis of microRNA156 and its targets, the genes encoding SQUAMOSA promoter-binding protein-like (SPL) transcription factors, in the grass family Poaceae
Abstract
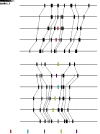
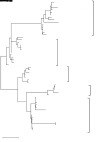
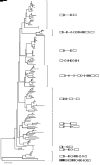
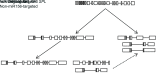
MicroRNAs (miRNAs) are endogenous small non-coding RNAs that play an important role in post-transcriptional gene regulation in plants and animals by targeting messenger RNAs (mRNAs) for cleavage or repressing translation of specific mRNAs. The first miRNA identified in plants, miRNA156 (miR156), targets the SQUAMOSA promoter-binding protein-like (SPL) transcription factors, which play critical roles in plant phase transition, flower and plant architecture, and fruit development. We identified multiple copies of MIR156 and SPL in the rice, Brachypodium, sorghum, maize, and foxtail millet genomes. Sequence and chromosomal synteny analysis showed that both MIR156s and SPLs are conserved across species in the grass family. Analysis of expression data of the SPLs in eleven juvenile and adult rice tissues revealed that four non-miR156-targeted genes were highly expressed and three miR156-targeted genes were only slightly expressed in all tissues/developmental stages. The remaining SPLs were highly expressed in the juvenile stage, but their expression was lower in the adult stage. It has been proposed that under strong selective pressure, non-miR156-targeted mRNA may be able to re-structure to form a miRNA-responsive element. In our analysis, some non-miR156-targeted SPLs (SPL5/8/10) had gene structure and gene expression patterns similar to those of miR156-targeted genes, suggesting that they could diversify into miR156-targeted genes. DNA methylation profiles of SPLs and MIR156s in different rice tissues showed diverse methylation patterns, and hypomethylation of non-CG sites was observed in rice endosperm. Our findings suggested that MIR156s and SPLs had different origination and evolutionary mechanisms: the SPLs appear to have resulted from vertical evolution, whereas MIR156s appear to have resulted from strong evolutionary selection on mature sequences.
Keywords: DNA methylation; Gene expression; Grass genome; MicroRNA156 (miR156); SQUAMOSA promoter-binding protein-like (SPL) gene.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

