Polyadenylation-related isoform switching in human evolution revealed by full-length transcript structure
- PMID: 33973996
- PMCID: PMC8574621
- DOI: 10.1093/bib/bbab157
Polyadenylation-related isoform switching in human evolution revealed by full-length transcript structure
Abstract
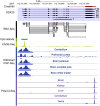
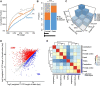
Rhesus macaque is a unique nonhuman primate model for human evolutionary and translational study, but the error-prone gene models critically limit its applications. Here, we de novo defined full-length macaque gene models based on single molecule, long-read transcriptome sequencing in four macaque tissues (frontal cortex, cerebellum, heart and testis). Overall, 8 588 227 poly(A)-bearing complementary DNA reads with a mean length of 14 106 nt were generated to compile the backbone of macaque transcripts, with the fine-scale structures further refined by RNA sequencing and cap analysis gene expression sequencing data. In total, 51 605 macaque gene models were accurately defined, covering 89.7% of macaque or 75.7% of human orthologous genes. Based on the full-length gene models, we performed a human-macaque comparative analysis on polyadenylation (PA) regulation. Using macaque and mouse as outgroup species, we identified 79 distal PA events newly originated in humans and found that the strengthening of the distal PA sites, rather than the weakening of the proximal sites, predominantly contributes to the origination of these human-specific isoforms. Notably, these isoforms are selectively constrained in general and contribute to the temporospatially specific reduction of gene expression, through the tinkering of previously existed mechanisms of nuclear retention and microRNA (miRNA) regulation. Overall, the protocol and resource highlight the application of bioinformatics in integrating multilayer genomics data to provide an intact reference for model animal studies, and the isoform switching detected may constitute a hitherto underestimated regulatory layer in shaping the human-specific transcriptome and phenotypic changes.
Keywords: Iso-Seq; gene models; polyadenylation; rhesus macaque; subcellular localization; transcriptome evolution.
© The Author(s) 2021. Published by Oxford University Press.
Figures




Similar articles
-
Isoform Evolution in Primates through Independent Combination of Alternative RNA Processing Events.Mol Biol Evol. 2017 Oct 1;34(10):2453-2468. doi: 10.1093/molbev/msx212. Mol Biol Evol. 2017. PMID: 28957512 Free PMC article.
-
A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing.BMC Genomics. 2017 May 22;18(1):395. doi: 10.1186/s12864-017-3757-8. BMC Genomics. 2017. PMID: 28532419 Free PMC article.
-
Evolutionary interrogation of human biology in well-annotated genomic framework of rhesus macaque.Mol Biol Evol. 2014 May;31(5):1309-24. doi: 10.1093/molbev/msu084. Epub 2014 Feb 27. Mol Biol Evol. 2014. PMID: 24577841 Free PMC article.
-
Analyses of alternative polyadenylation: from old school biochemistry to high-throughput technologies.BMB Rep. 2017 Apr;50(4):201-207. doi: 10.5483/bmbrep.2017.50.4.019. BMB Rep. 2017. PMID: 28148393 Free PMC article. Review.
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
-
De novo genes with an lncRNA origin encode unique human brain developmental functionality.Nat Ecol Evol. 2023 Feb;7(2):264-278. doi: 10.1038/s41559-022-01925-6. Epub 2023 Jan 2. Nat Ecol Evol. 2023. PMID: 36593289 Free PMC article.
-
Comparative transcriptome analysis between rhesus macaques ( Macaca mulatta) and crab-eating macaques ( M. fascicularis).Zool Res. 2024 Mar 18;45(2):299-310. doi: 10.24272/j.issn.2095-8137.2023.322. Zool Res. 2024. PMID: 38485500 Free PMC article.
References
-
- Rhesus Macaque Genome Sequencing and Analysis Consortium, Gibbs RA, Rogers J, et al. . Evolutionary and biomedical insights from the rhesus macaque genome. Science 2007;316:222–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

