(CO2)n+, (H2O)n+, and (H2O)n+ (CO2) gas cluster ion beam secondary ion mass spectrometry: analysis of lipid extracts, cells, and Alzheimer's model mouse brain tissue
- PMID: 33974088
- PMCID: PMC8222020
- DOI: 10.1007/s00216-021-03372-x
(CO2)n+, (H2O)n+, and (H2O)n+ (CO2) gas cluster ion beam secondary ion mass spectrometry: analysis of lipid extracts, cells, and Alzheimer's model mouse brain tissue
Abstract
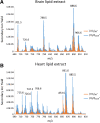
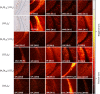
This work assesses the potential of new water cluster-based ion beams for improving the capabilities of secondary ion mass spectrometry (SIMS) for in situ lipidomics. The effect of water clusters was compared to carbon dioxide clusters, along with the effect of using pure water clusters compared to mixed water and carbon dioxide clusters. A signal increase was found when using pure water clusters. However, when analyzing cells, a more substantial signal increase was found in positive ion mode when the water clusters also contained carbon dioxide, suggesting that additional reactions are in play. The effects of using a water primary ion beam on a more complex sample were investigated by analyzing brain tissue from an Alzheimer's disease transgenic mouse model. The results indicate that the ToF-SIMS results are approaching those from MALDI as ToF-SIMS was able to image lyso-phosphocholine (LPC) lipids, a lipid class that for a long time has eluded detection during SIMS analyses. Gangliosides, sulfatides, and cholesterol were also imaged.
Keywords: Alzheimer’s; Imaging; Lipids; Mass spectrometry; SIMS; Water clusters.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Dowsett D, Wirtz T. Co-registered in situ secondary electron and mass spectral imaging on the helium ion microscope demonstrated using lithium titanate and magnesium oxide nanoparticles. Anal Chem. 2017;89(17):8957–8965. - PubMed
-
- Vickerman JC. Molecular imaging and depth profiling by mass spectrometry-SIMS, MALDI or DESI? Analyst. 2011;136(11):2199–2217. - PubMed
-
- Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. - PubMed
-
- Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

