Integrated profiling identifies ITGB3BP as prognostic biomarker for hepatocellular carcinoma
- PMID: 33974527
- PMCID: PMC8554696
- DOI: 10.17305/bjbms.2021.5690
Integrated profiling identifies ITGB3BP as prognostic biomarker for hepatocellular carcinoma
Abstract
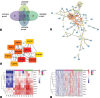
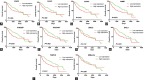
Hepatocellular carcinoma (HCC) is a highly malignant tumor. In this study, we sought to identify a novel biomarker for HCC by analyzing transcriptome and clinical data. The R software was used to analyze the differentially expressed genes (DEGs) in the datasets GSE74656 and GSE84598 downloaded from the Gene Expression Omnibus database, followed by a functional annotation. A total of 138 shared DEGs were screened from two datasets. They were mainly enriched in the "Metabolic pathways" pathway (Padj = 8.21E-08) and involved in the carboxylic acid metabolic process (Padj = 0.0004). The top 10 hub genes were found by protein-protein interaction analysis and were upregulated in HCC tissues compared to normal tissues in The Cancer Genome Atlas database. Survival analysis distinguished 8 hub genes CENPE, SPDL1, Hyaluronan-mediated motility receptor, Rac GTPase activating protein 1, Thyroid hormone receptor interactor 13, cytoskeleton-associated protein (CKAP) 2, CKAP5, and Integrin subunit beta 3 binding protein (ITGB3BP) were considered as prognostic hub genes. Multivariate cox regression analysis indicated that all the prognostic hub genes were independent prognostic factors for HCC. Furthermore, the receiver operating characteristic curve revealed that the 8-hub genes model had better prediction performance for overall survival compared to the T stage (p = 0.008) and significantly improved the prediction value of the T stage (p = 0.002). The Human Protein Atlas showed that the protein expression of ITGB3BP was upregulated in HCC, so the expression of ITGB3BP was further verified in our cohort. The results showed that ITGB3BP was upregulated in HCC tissues and was significantly associated with lymph node metastasis.
Conflict of interest statement
Conflict of interest statement: The authors declare no conflict of interests
Figures






Similar articles
-
Identification of Potential Hub Genes Related to Diagnosis and Prognosis of Hepatitis B Virus-Related Hepatocellular Carcinoma via Integrated Bioinformatics Analysis.Biomed Res Int. 2020 Dec 8;2020:4251761. doi: 10.1155/2020/4251761. eCollection 2020. Biomed Res Int. 2020. PMID: 33376723 Free PMC article.
-
Identifying hepatocellular carcinoma-related hub genes by bioinformatics analysis and CYP2C8 is a potential prognostic biomarker.Gene. 2019 May 25;698:9-18. doi: 10.1016/j.gene.2019.02.062. Epub 2019 Feb 27. Gene. 2019. PMID: 30825595
-
Bioinformatics Analysis of Candidate Genes and Pathways Related to Hepatocellular Carcinoma in China: A Study Based on Public Databases.Pathol Oncol Res. 2021 Mar 26;27:588532. doi: 10.3389/pore.2021.588532. eCollection 2021. Pathol Oncol Res. 2021. PMID: 34257537 Free PMC article.
-
Screening Hub Genes as Prognostic Biomarkers of Hepatocellular Carcinoma by Bioinformatics Analysis.Cell Transplant. 2019 Dec;28(1_suppl):76S-86S. doi: 10.1177/0963689719893950. Epub 2019 Dec 11. Cell Transplant. 2019. PMID: 31822116 Free PMC article.
-
Identification of Hub Genes and Analysis of Prognostic Values in Hepatocellular Carcinoma by Bioinformatics Analysis.Am J Med Sci. 2020 Apr;359(4):226-234. doi: 10.1016/j.amjms.2020.01.009. Epub 2020 Jan 21. Am J Med Sci. 2020. PMID: 32200915
Cited by
-
Differential methylation patterns in lean and obese non-alcoholic steatohepatitis-associated hepatocellular carcinoma.BMC Cancer. 2022 Dec 6;22(1):1276. doi: 10.1186/s12885-022-10389-7. BMC Cancer. 2022. PMID: 36474183 Free PMC article.
-
LncRNA NR_045147 modulates osteogenic differentiation and migration in PDLSCs via ITGB3BP degradation and mitochondrial dysfunction.Stem Cells Transl Med. 2025 Feb 11;14(2):szae088. doi: 10.1093/stcltm/szae088. Stem Cells Transl Med. 2025. PMID: 39674578 Free PMC article.
-
Kinesin-7 CENP-E in tumorigenesis: Chromosome instability, spindle assembly checkpoint, and applications.Front Mol Biosci. 2024 Mar 15;11:1366113. doi: 10.3389/fmolb.2024.1366113. eCollection 2024. Front Mol Biosci. 2024. PMID: 38560520 Free PMC article. Review.
References
-
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–14. https://doi.org/10.1016/s0140-6736(18)30010-2. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492. - PubMed
-
- Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, et al. Global patterns of hepatocellular carcinoma management from diagnosis to death:The BRIDGE Study. Liver Int. 2015;35(9):2155–66. https://doi.org/10.1111/liv.12818. - PMC - PubMed
-
- Singal AG, Parikh ND, Rich NE, John BV, Pillai A. Hoshida Y, editor. Hepatocellular Carcinoma Surveillance and Staging. Hepatocellular Carcinoma:Translational Precision Medicine Approaches. Cham CH:Springer Nature Switzerland AG. 2019:27–51. https://doi.org/10.1007/978-3-030-21540-8_2.
-
- Xue R, Li R, Guo H, Guo L, Su Z, Ni X, et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology. 2016;150(4):998–1008. https://doi.org/10.1053/j.gastro.2015.12.033. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

