Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities
- PMID: 33974846
- PMCID: PMC8078906
- DOI: 10.1016/j.celrep.2021.109126
Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities
Abstract
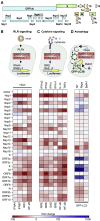
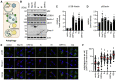
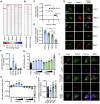
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) evades most innate immune responses but may still be vulnerable to some. Here, we systematically analyze the impact of SARS-CoV-2 proteins on interferon (IFN) responses and autophagy. We show that SARS-CoV-2 proteins synergize to counteract anti-viral immune responses. For example, Nsp14 targets the type I IFN receptor for lysosomal degradation, ORF3a prevents fusion of autophagosomes and lysosomes, and ORF7a interferes with autophagosome acidification. Most activities are evolutionarily conserved. However, SARS-CoV-2 Nsp15 antagonizes IFN signaling less efficiently than the orthologs of closely related RaTG13-CoV and SARS-CoV-1. Overall, SARS-CoV-2 proteins counteract autophagy and type I IFN more efficiently than type II or III IFN signaling, and infection experiments confirm potent inhibition by IFN-γ and -λ1. Our results define the repertoire and selected mechanisms of SARS-CoV-2 innate immune antagonists but also reveal vulnerability to type II and III IFN that may help to develop safe and effective anti-viral approaches.
Keywords: COVID-19; SARS-CoV; SARS-CoV-2; autophagy; cytokine; immune evasion; innate immunity; interferon.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests F.I.S. is a co-founder of DiosCURE Therapeutics SE and a consultant to IFM Therapeutics.
Figures

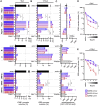
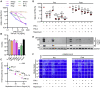
Comment in
-
Manipulation of autophagy by SARS-CoV-2 proteins.Autophagy. 2021 Sep;17(9):2659-2661. doi: 10.1080/15548627.2021.1953847. Epub 2021 Jul 19. Autophagy. 2021. PMID: 34281462 Free PMC article.
References
-
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:80. - PMC - PubMed
-
- Bozzo C.P., Nchioua R., Volcic M., Wettstein L., Weil T., Krüger J., Heller S., Conzelmann C., Müller J., Gross R. IFITM proteins promote SARS-CoV-2 infection of human lung cells. bioRxiv. 2020 doi: 10.1101/2020.08.18.255935. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

