The DnaK/DnaJ Chaperone System Enables RNA Polymerase-DksA Complex Formation in Salmonella Experiencing Oxidative Stress
- PMID: 33975942
- PMCID: PMC8262869
- DOI: 10.1128/mBio.03443-20
The DnaK/DnaJ Chaperone System Enables RNA Polymerase-DksA Complex Formation in Salmonella Experiencing Oxidative Stress
Abstract
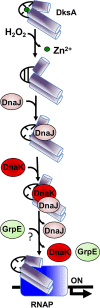
Our previous biochemical approaches showed that the oxidoreductase activity of the DnaJ protein facilitates the interaction of oxidized DksA with RNA polymerase. Investigations herein demonstrate that under biologically relevant conditions the DnaJ- and DksA-codependent activation of the stringent response in Salmonella undergoing oxidative stress involves the DnaK chaperone. Oxidation of DksA cysteine residues stimulates redox-based and holdase interactions with zinc-binding and C-terminal domains of DnaJ. Genetic and biochemical evidence indicates that His33 in the HPD motif in the J domain of DnaJ facilitates interactions of unfolded DksA with DnaK. A mutation in His33 in the J domain prevents the presentation of unfolded DksA to DnaK without limiting the oxidoreductase activity mapped to DnaJ's zinc-2 site. Thr199 in the ATPase catalytic site of DnaK is required for the formation of the DksA/RNA polymerase complex. The DnaK/DnaJ/DksA complex enables the formation of an enzymatically active RNA polymerase holoenzyme that stimulates transcription of branched-chain amino acid and histidine metabolic genes in Salmonella exposed to reactive oxygen species. The DnaK/DnaJ chaperone protects Salmonella against the cytotoxicity associated with reactive oxygen species generated by the phagocyte NADPH oxidase in the innate host response. The antioxidant defenses associated with DnaK/DnaJ can in part be ascribed to the elicitation of the DksA-dependent stringent response and the protection this chaperone system provides against protein carbonylation in Salmonella undergoing oxidative stress.IMPORTANCE DksA was discovered 30 years ago in a screen for suppressors that reversed the thermosensitivity of Escherichia coli mutant strains deficient in DnaK/DnaJ, raising the possibility that this chaperone system may control DksA function. Since its serendipitous discovery, DksA has emerged as a key activator of the transcriptional program called the stringent response in Gram-negative bacteria experiencing diverse adverse conditions, including nutritional starvation or oxidative stress. DksA activates the stringent response through the allosteric control this regulatory protein exerts on the kinetics of RNA polymerase promoter open complexes. Recent investigations have shown that DksA overexpression protects dnaKJ mutant bacteria against heat shock indirectly via the ancestral chaperone polyphosphate, casting doubt on a possible complexation of DnaK, DnaJ, and DksA. Nonetheless, research presented herein demonstrates that the cochaperones DnaK and DnaJ enable DksA/RNA polymerase complex formation in response to oxidative stress.
Keywords: DksA; DnaJ; DnaK; Salmonella Typhimurium; chaperone; hydrogen peroxide; oxidative stress; redox; stringent response.
Copyright © 2021 Kim et al.
Figures






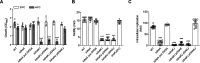
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
