Activation of transmembrane receptor tyrosine kinase DDR1-STAT3 cascade by extracellular matrix remodeling promotes liver metastatic colonization in uveal melanoma
- PMID: 33976105
- PMCID: PMC8113510
- DOI: 10.1038/s41392-021-00563-x
Activation of transmembrane receptor tyrosine kinase DDR1-STAT3 cascade by extracellular matrix remodeling promotes liver metastatic colonization in uveal melanoma
Abstract
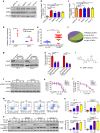
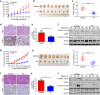
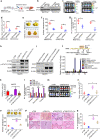
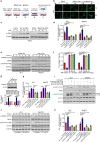
Colonization is believed a rate-limiting step of metastasis cascade. However, its underlying mechanism is not well understood. Uveal melanoma (UM), which is featured with single organ liver metastasis, may provide a simplified model for realizing the complicated colonization process. Because DDR1 was identified to be overexpressed in UM cell lines and specimens, and abundant pathological deposition of extracellular matrix collagen, a type of DDR1 ligand, was noted in the microenvironment of liver in metastatic patients with UM, we postulated the hypothesis that DDR1 and its ligand might ignite the interaction between UM cells and their surrounding niche of liver thereby conferring strengthened survival, proliferation, stemness and eventually promoting metastatic colonization in liver. We tested this hypothesis and found that DDR1 promoted these malignant cellular phenotypes and facilitated metastatic colonization of UM in liver. Mechanistically, UM cells secreted TGF-β1 which induced quiescent hepatic stellate cells (qHSCs) into activated HSCs (aHSCs) which secreted collagen type I. Such a remodeling of extracellular matrix, in turn, activated DDR1, strengthening survival through upregulating STAT3-dependent Mcl-1 expression, enhancing stemness via upregulating STAT3-dependent SOX2, and promoting clonogenicity in cancer cells. Targeting DDR1 by using 7rh, a specific inhibitor, repressed proliferation and survival in vitro and in vivo outgrowth. More importantly, targeting cancer cells by pharmacological inactivation of DDR1 or targeting microenvironmental TGF-β1-collagen I loop exhibited a prominent anti-metastasis effect in mice. In conclusion, targeting DDR1 signaling and TGF-β signaling may be a novel approach to diminish hepatic metastasis in UM.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Neuropeptide Precursor VGF Promotes Liver Metastatic Colonization of Gαq Mutant Uveal Melanoma by Facilitating Tumor Microenvironment via Paracrine Loops.Adv Sci (Weinh). 2024 Dec;11(46):e2407967. doi: 10.1002/advs.202407967. Epub 2024 Oct 18. Adv Sci (Weinh). 2024. PMID: 39422674 Free PMC article.
-
Transcriptional inhibition by CDK7/9 inhibitor SNS-032 abrogates oncogene addiction and reduces liver metastasis in uveal melanoma.Mol Cancer. 2019 Sep 16;18(1):140. doi: 10.1186/s12943-019-1070-7. Mol Cancer. 2019. PMID: 31526394 Free PMC article.
-
Verification of EZH2 as a druggable target in metastatic uveal melanoma.Mol Cancer. 2020 Mar 4;19(1):52. doi: 10.1186/s12943-020-01173-x. Mol Cancer. 2020. PMID: 32127003 Free PMC article.
-
Uveal melanoma pathobiology: Metastasis to the liver.Semin Cancer Biol. 2021 Jun;71:65-85. doi: 10.1016/j.semcancer.2020.05.003. Epub 2020 May 22. Semin Cancer Biol. 2021. PMID: 32450140 Review.
-
Signaling by discoidin domain receptor 1 in cancer metastasis.Cell Adh Migr. 2018;12(4):315-323. doi: 10.1080/19336918.2018.1520556. Epub 2018 Oct 13. Cell Adh Migr. 2018. PMID: 30187813 Free PMC article. Review.
Cited by
-
[The Role and Significance of Hepatic Environmental Cells in Tumor Metastatic Colonization to Liver].Sichuan Da Xue Xue Bao Yi Xue Ban. 2023 May;54(3):469-474. doi: 10.12182/20230560301. Sichuan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37248570 Free PMC article. Review. Chinese.
-
Emerging strategies and translational advancements of DDR1 in oncology.Discov Oncol. 2025 Mar 30;16(1):428. doi: 10.1007/s12672-025-02107-z. Discov Oncol. 2025. PMID: 40159417 Free PMC article. Review.
-
Effects of solamargine in hepatic metastasis of colorectal cancer: induction of ferroptosis and elimination of cancer stem cells.Chin Med. 2025 Jul 11;20(1):110. doi: 10.1186/s13020-025-01171-5. Chin Med. 2025. PMID: 40646619 Free PMC article.
-
Neuropeptide Precursor VGF Promotes Liver Metastatic Colonization of Gαq Mutant Uveal Melanoma by Facilitating Tumor Microenvironment via Paracrine Loops.Adv Sci (Weinh). 2024 Dec;11(46):e2407967. doi: 10.1002/advs.202407967. Epub 2024 Oct 18. Adv Sci (Weinh). 2024. PMID: 39422674 Free PMC article.
-
Identification of hub genes and pathways in lung metastatic colorectal cancer.BMC Cancer. 2023 Apr 6;23(1):323. doi: 10.1186/s12885-023-10792-8. BMC Cancer. 2023. PMID: 37024866 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

