FUT8-mediated aberrant N-glycosylation of B7H3 suppresses the immune response in triple-negative breast cancer
- PMID: 33976130
- PMCID: PMC8113546
- DOI: 10.1038/s41467-021-22618-x
FUT8-mediated aberrant N-glycosylation of B7H3 suppresses the immune response in triple-negative breast cancer
Abstract
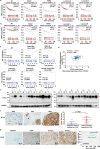
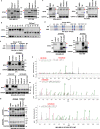
Most patients with triple negative breast cancer (TNBC) do not respond to anti-PD1/PDL1 immunotherapy, indicating the necessity to explore immune checkpoint targets. B7H3 is a highly glycosylated protein. However, the mechanisms of B7H3 glycosylation regulation and whether the sugar moiety contributes to immunosuppression are unclear. Here, we identify aberrant B7H3 glycosylation and show that N-glycosylation of B7H3 at NXT motif sites is responsible for its protein stability and immunosuppression in TNBC tumors. The fucosyltransferase FUT8 catalyzes B7H3 core fucosylation at N-glycans to maintain its high expression. Knockdown of FUT8 rescues glycosylated B7H3-mediated immunosuppressive function in TNBC cells. Abnormal B7H3 glycosylation mediated by FUT8 overexpression can be physiologically important and clinically relevant in patients with TNBC. Notably, the combination of core fucosylation inhibitor 2F-Fuc and anti-PDL1 results in enhanced therapeutic efficacy in B7H3-positive TNBC tumors. These findings suggest that targeting the FUT8-B7H3 axis might be a promising strategy for improving anti-tumor immune responses in patients with TNBC.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
FUT8 Is a Critical Driver of Prostate Tumour Growth and Can Be Targeted Using Fucosylation Inhibitors.Cancer Med. 2025 May;14(10):e70959. doi: 10.1002/cam4.70959. Cancer Med. 2025. PMID: 40387385 Free PMC article.
-
Targeting site-specific N-glycosylated B7H3 induces potent antitumor immunity.Nat Commun. 2025 Apr 14;16(1):3546. doi: 10.1038/s41467-025-58740-3. Nat Commun. 2025. PMID: 40229277 Free PMC article.
-
Mammalian α-1,6-Fucosyltransferase (FUT8) Is the Sole Enzyme Responsible for the N-Acetylglucosaminyltransferase I-independent Core Fucosylation of High-mannose N-Glycans.J Biol Chem. 2016 May 20;291(21):11064-71. doi: 10.1074/jbc.M116.720789. Epub 2016 Mar 23. J Biol Chem. 2016. PMID: 27008861 Free PMC article.
-
FUT8 Alpha-(1,6)-Fucosyltransferase in Cancer.Int J Mol Sci. 2021 Jan 5;22(1):455. doi: 10.3390/ijms22010455. Int J Mol Sci. 2021. PMID: 33466384 Free PMC article. Review.
-
Distinctive domains and activity regulation of core fucosylation enzyme FUT8.Biochim Biophys Acta Gen Subj. 2024 Apr;1868(4):130561. doi: 10.1016/j.bbagen.2024.130561. Epub 2024 Jan 12. Biochim Biophys Acta Gen Subj. 2024. PMID: 38218458 Review.
Cited by
-
Nanomaterial-assisted CRISPR gene-engineering - A hallmark for triple-negative breast cancer therapeutics advancement.Mater Today Bio. 2022 Oct 4;16:100450. doi: 10.1016/j.mtbio.2022.100450. eCollection 2022 Dec. Mater Today Bio. 2022. PMID: 36267139 Free PMC article. Review.
-
The Multifaceted Role of FUT8 in Tumorigenesis: From Pathways to Potential Clinical Applications.Int J Mol Sci. 2024 Jan 15;25(2):1068. doi: 10.3390/ijms25021068. Int J Mol Sci. 2024. PMID: 38256141 Free PMC article. Review.
-
FUT8 is regulated by miR-122-5p and promotes malignancies in intrahepatic cholangiocarcinoma via PI3K/AKT signaling.Cell Oncol (Dordr). 2023 Feb;46(1):79-91. doi: 10.1007/s13402-022-00736-y. Epub 2022 Nov 8. Cell Oncol (Dordr). 2023. PMID: 36348252
-
CD276 regulates the immune escape of esophageal squamous cell carcinoma through CXCL1-CXCR2 induced NETs.J Immunother Cancer. 2024 May 9;12(5):e008662. doi: 10.1136/jitc-2023-008662. J Immunother Cancer. 2024. PMID: 38724465 Free PMC article.
-
Post-translational modifications of immune checkpoints: unlocking new potentials in cancer immunotherapy.Exp Hematol Oncol. 2025 Mar 14;14(1):37. doi: 10.1186/s40164-025-00627-6. Exp Hematol Oncol. 2025. PMID: 40087690 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

