Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities
- PMID: 33976161
- PMCID: PMC8113577
- DOI: 10.1038/s41467-021-22849-y
Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities
Abstract
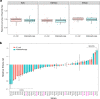
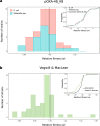
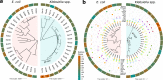
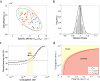
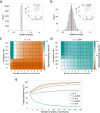
Plasmid persistence in bacterial populations is strongly influenced by the fitness effects associated with plasmid carriage. However, plasmid fitness effects in wild-type bacterial hosts remain largely unexplored. In this study, we determined the fitness effects of the major antibiotic resistance plasmid pOXA-48_K8 in wild-type, ecologically compatible enterobacterial isolates from the human gut microbiota. Our results show that although pOXA-48_K8 produced an overall reduction in bacterial fitness, it produced small effects in most bacterial hosts, and even beneficial effects in several isolates. Moreover, genomic results showed a link between pOXA-48_K8 fitness effects and bacterial phylogeny, helping to explain plasmid epidemiology. Incorporating our fitness results into a simple population dynamics model revealed a new set of conditions for plasmid stability in bacterial communities, with plasmid persistence increasing with bacterial diversity and becoming less dependent on conjugation. These results help to explain the high prevalence of plasmids in the greatly diverse natural microbial communities.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- San Millan, A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol.26, 978–985 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

