Taxonomic signatures of cause-specific mortality risk in human gut microbiome
- PMID: 33976176
- PMCID: PMC8113604
- DOI: 10.1038/s41467-021-22962-y
Taxonomic signatures of cause-specific mortality risk in human gut microbiome
Abstract
The collection of fecal material and developments in sequencing technologies have enabled standardised and non-invasive gut microbiome profiling. Microbiome composition from several large cohorts have been cross-sectionally linked to various lifestyle factors and diseases. In spite of these advances, prospective associations between microbiome composition and health have remained uncharacterised due to the lack of sufficiently large and representative population cohorts with comprehensive follow-up data. Here, we analyse the long-term association between gut microbiome variation and mortality in a well-phenotyped and representative population cohort from Finland (n = 7211). We report robust taxonomic and functional microbiome signatures related to the Enterobacteriaceae family that are associated with mortality risk during a 15-year follow-up. Our results extend previous cross-sectional studies, and help to establish the basis for examining long-term associations between human gut microbiome composition, incident outcomes, and general health status.
Conflict of interest statement
The authors declare the following competing interests: V.S. has consulted for Novo Nordisk and Sanofi and received honoraria from these companies. He also has ongoing research collaboration with Bayer AG, all unrelated to this study. The other authors declare no competing interests.
Figures

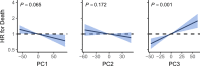

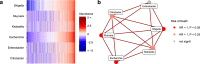
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

