N-aryl pyrido cyanine derivatives are nuclear and organelle DNA markers for two-photon and super-resolution imaging
- PMID: 33976192
- PMCID: PMC8113587
- DOI: 10.1038/s41467-021-23019-w
N-aryl pyrido cyanine derivatives are nuclear and organelle DNA markers for two-photon and super-resolution imaging
Abstract
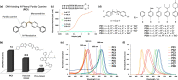
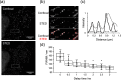
Live cell imaging using fluorescent DNA markers are an indispensable molecular tool in various biological and biomedical fields. It is a challenge to develop DNA probes that avoid UV light photo-excitation, have high specificity for DNA, are cell-permeable and are compatible with cutting-edge imaging techniques such as super-resolution microscopy. Herein, we present N-aryl pyrido cyanine (N-aryl-PC) derivatives as a class of long absorption DNA markers with absorption in the wide range of visible light. The high DNA specificity and membrane permeability allow the staining of both organelle DNA as well as nuclear DNA, in various cell types, including plant tissues, without the need for washing post-staining. N-aryl-PC dyes are also highly compatible with a separation of photon by lifetime tuning method in stimulated emission depletion microscopy (SPLIT-STED) for super-resolution imaging as well as two-photon microscopy for deep tissue imaging, making it a powerful tool in the life sciences.
Conflict of interest statement
The patent application, “Cyanine compound and fluorescent dye” (WO2019012963) invented by Y.S. and K.U. has been published (
Figures


References
-
- Tiselius A. A new apparatus for electrophoretic analysis of colloidal mixtures. Trans. Faraday Soc. 1937;33:524–531. doi: 10.1039/tf9373300524. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

