Parameter-free rendering of single-molecule localization microscopy data for parameter-free resolution estimation
- PMID: 33976358
- PMCID: PMC8113488
- DOI: 10.1038/s42003-021-02086-1
Parameter-free rendering of single-molecule localization microscopy data for parameter-free resolution estimation
Abstract
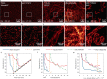
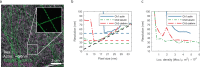
Localization microscopy is a super-resolution imaging technique that relies on the spatial and temporal separation of blinking fluorescent emitters. These blinking events can be individually localized with a precision significantly smaller than the classical diffraction limit. This sub-diffraction localization precision is theoretically bounded by the number of photons emitted per molecule and by the sensor noise. These parameters can be estimated from the raw images. Alternatively, the resolution can be estimated from a rendered image of the localizations. Here, we show how the rendering of localization datasets can influence the resolution estimation based on decorrelation analysis. We demonstrate that a modified histogram rendering, termed bilinear histogram, circumvents the biases introduced by Gaussian or standard histogram rendering. We propose a parameter-free processing pipeline and show that the resolution estimation becomes a function of the localization density and the localization precision, on both simulated and state-of-the-art experimental datasets.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

