Randomized prospective evaluation of genome sequencing versus standard-of-care as a first molecular diagnostic test
- PMID: 33976420
- PMCID: PMC8488861
- DOI: 10.1038/s41436-021-01193-y
Randomized prospective evaluation of genome sequencing versus standard-of-care as a first molecular diagnostic test
Abstract
Purpose: To evaluate the diagnostic yield and clinical relevance of clinical genome sequencing (cGS) as a first genetic test for patients with suspected monogenic disorders.
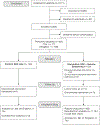
Methods: We conducted a prospective randomized study with pediatric and adult patients recruited from genetics clinics at Massachusetts General Hospital who were undergoing planned genetic testing. Participants were randomized into two groups: standard-of-care genetic testing (SOC) only or SOC and cGS.
Results: Two hundred four participants were enrolled, 202 were randomized to one of the intervention arms, and 99 received cGS. In total, cGS returned 16 molecular diagnoses that fully or partially explained the indication for testing in 16 individuals (16.2% of the cohort, 95% confidence interval [CI] 8.9-23.4%), which was not significantly different from SOC (18.2%, 95% CI 10.6-25.8%, P = 0.71). An additional eight molecular diagnoses reported by cGS had uncertain relevance to the participant's phenotype. Nevertheless, referring providers considered 20/24 total cGS molecular diagnoses (83%) to be explanatory for clinical features or worthy of additional workup.
Conclusion: cGS is technically suitable as a first genetic test. In our cohort, diagnostic yield was not significantly different from SOC. Further studies addressing other variant types and implementation challenges are needed to support feasibility and utility of broad-scale cGS adoption.
© 2021. The Author(s), under exclusive licence to the American College of Medical Genetics and Genomics.
Figures


References
-
- Ross LF, Saal HM, David KL & Anderson RR Technical report: ethical and policy issues in genetic testing and screening of children. Genet. Med 15, 234–245 (2013). - PubMed
-
- National Comprehensive Cancer Network. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic (Version 1.2020). https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf (2020). - PubMed
-
- Xue Y, Ankala A, Wilcox WR & Hegde MR Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet. Med 17, 444–451 (2015). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

