Gut microbiome of migratory shorebirds: Current status and future perspectives
- PMID: 33976772
- PMCID: PMC8093701
- DOI: 10.1002/ece3.7390
Gut microbiome of migratory shorebirds: Current status and future perspectives
Abstract
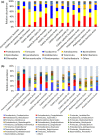
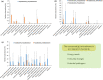
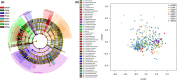
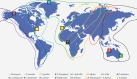
Migratory shorebirds have many unique life history characteristics, such as long-distance travel between breeding sites, stopover sites, and wintering sites. The physiological challenges for migrant energy requirement and immunity may affect their gut microbiome community. Here, we reviewed the specific features (e.g., relatively high proportion of Corynebacterium and Fusobacterium) in the gut microbiome of 18 migratory shorebirds, and the factors (e.g., diet, migration, environment, and phylogeny) affecting the gut microbiome. We discussed possible future studies of the gut microbiome in migratory shorebirds, including the composition and function of the spatial-temporal gut microbiome, and the potential contributions made by the gut microbiome to energy requirement during migration.
Keywords: composition and function; energy requirement; migratory shorebirds; the gut microbiome.
© 2021 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declared no conflicts of interest relevant to this manuscript.
Figures

References
-
- Alerstam, T. , Hedenström, A. , & Åkesson, S. (2003). Long‐distance migration: Evolution and determinants. Oikos, 103, 247–260. 10.1034/j.1600-0706.2003.12559.x - DOI
-
- Archie, E. A. , & Tung, J. (2015). Social behavior and the microbiome. Current Opinion in Behavioral Sciences, 6, 28–34. 10.1016/j.cobeha.2015.07.008 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

