Cell size, genome size, and maximum growth rate are near-independent dimensions of ecological variation across bacteria and archaea
- PMID: 33976787
- PMCID: PMC8093753
- DOI: 10.1002/ece3.7290
Cell size, genome size, and maximum growth rate are near-independent dimensions of ecological variation across bacteria and archaea
Abstract
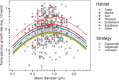
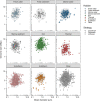
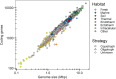
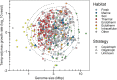
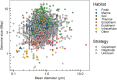
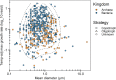
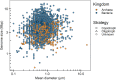
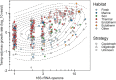
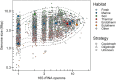
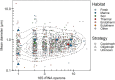
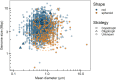
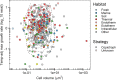
Among bacteria and archaea, maximum relative growth rate, cell diameter, and genome size are widely regarded as important influences on ecological strategy. Via the most extensive data compilation so far for these traits across all clades and habitats, we ask whether they are correlated and if so how. Overall, we found little correlation among them, indicating they should be considered as independent dimensions of ecological variation. Nor was correlation evident within particular habitat types. A weak nonlinearity (6% of variance) was found whereby high maximum growth rates (temperature-adjusted) tended to occur in the midrange of cell diameters. Species identified in the literature as oligotrophs or copiotrophs were clearly separated on the dimension of maximum growth rate, but not on the dimensions of genome size or cell diameter.
Keywords: archaea; bacteria; cell diameter; ecological strategies; genome size; maximum growth rate; traits.
© 2021 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
None of the authors have any conflict of interest.
Figures




References
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

