Diversity and circulation of Jingmen tick virus in ticks and mammals
- PMID: 33976906
- PMCID: PMC8097133
- DOI: 10.1093/ve/veaa051
Diversity and circulation of Jingmen tick virus in ticks and mammals
Abstract
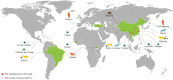
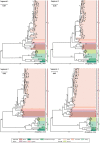
Since its initial identification in ticks in 2010, Jingmen tick virus (JMTV) has been described in cattle, rodents, and primates. To better understand the diversity, evolution, and transmission of JMTV, we sampled 215 ticks, 104 cattle bloods, 216 bats, and 119 rodents in Wenzhou city, Zhejiang Province, China as well as 240 bats from Guizhou and Henan Provinces. JMTV was identified in 107 ticks (from two species), 54 bats (eleven species), 8 rodents (three species), and 10 cattle, with prevalence levels of 49.8, 11.8, 6.7, and 9.6 per cent, respectively, suggesting that bats may be a natural reservoir of JMTV. Phylogenetic analyses revealed that all the newly identified JMTVs were closely related to each other and to previously described viruses. Additionally, all tick and mammalian JMTV sampled in Wenzhou shared a consistent genomic structure, suggesting that the virus can cocirculate between ticks and mammals without observable variation in genome organization. All JMTVs sampled globally could be divided into two phylogenetic groups, Mantel tests suggested that geographic isolation, rather than host species, may be the main driver of JMTV diversity. However, the exact geographical origin of JMTV was difficult to determine, suggesting that this virus has a complex evolutionary history.
Keywords: Jingmen tick virus; diversity; evolution; mammals; ticks; transmission.
© The Author(s) 2020. Published by Oxford University Press.
Figures

Similar articles
-
Identification of Jingmen tick virus (JMTV) in Amblyomma testudinarium from Fujian Province, southeastern China.Parasit Vectors. 2022 Sep 27;15(1):339. doi: 10.1186/s13071-022-05478-2. Parasit Vectors. 2022. PMID: 36167570 Free PMC article.
-
Identification and characterization of Jingmen tick virus in rodents from Xinjiang, China.Infect Genet Evol. 2020 Oct;84:104411. doi: 10.1016/j.meegid.2020.104411. Epub 2020 Jun 10. Infect Genet Evol. 2020. PMID: 32531517 Free PMC article.
-
Identification and phylogenetic analysis of Jingmen tick virus in Jiangxi Province, China.Front Vet Sci. 2024 May 2;11:1375852. doi: 10.3389/fvets.2024.1375852. eCollection 2024. Front Vet Sci. 2024. PMID: 38756509 Free PMC article.
-
Jingmen tick virus: an emerging arbovirus with a global threat.mSphere. 2023 Oct 24;8(5):e0028123. doi: 10.1128/msphere.00281-23. Epub 2023 Sep 13. mSphere. 2023. PMID: 37702505 Free PMC article. Review.
-
The Ecology of New Constituents of the Tick Virome and Their Relevance to Public Health.Viruses. 2019 Jun 7;11(6):529. doi: 10.3390/v11060529. Viruses. 2019. PMID: 31181599 Free PMC article. Review.
Cited by
-
Identification of Jingmen tick virus (JMTV) in Amblyomma testudinarium from Fujian Province, southeastern China.Parasit Vectors. 2022 Sep 27;15(1):339. doi: 10.1186/s13071-022-05478-2. Parasit Vectors. 2022. PMID: 36167570 Free PMC article.
-
Geographical and Tick-Dependent Distribution of Flavi-Like Alongshan and Yanggou Tick Viruses in Russia.Viruses. 2021 Mar 11;13(3):458. doi: 10.3390/v13030458. Viruses. 2021. PMID: 33799742 Free PMC article.
-
Identification and phylogenetic analysis of Jingmen tick virus in ticks and sheep from Henan Province, China.Virol J. 2024 Dec 20;21(1):325. doi: 10.1186/s12985-024-02587-5. Virol J. 2024. PMID: 39707432 Free PMC article.
-
Identification of a putative new virus from the Jingmenvirus group in ticks from wild animals in Brazil.Virus Evol. 2025 Jun 27;11(1):veaf045. doi: 10.1093/ve/veaf045. eCollection 2025. Virus Evol. 2025. PMID: 40666309 Free PMC article.
-
A Novel Subtype of Bovine Hepacivirus Identified in Ticks Reveals the Genetic Diversity and Evolution of Bovine Hepacivirus.Viruses. 2021 Nov 2;13(11):2206. doi: 10.3390/v13112206. Viruses. 2021. PMID: 34835012 Free PMC article.
References
-
- Almagro Armenteros J. J. et al. (2019) ‘SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks’, Nature Biotechnology, 37: 420–3. - PubMed
LinkOut - more resources
Full Text Sources

