Alternation between taxonomically divergent hosts is not the major determinant of flavivirus evolution
- PMID: 33976907
- PMCID: PMC8093920
- DOI: 10.1093/ve/veab040
Alternation between taxonomically divergent hosts is not the major determinant of flavivirus evolution
Abstract
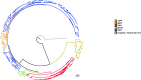
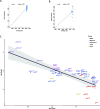
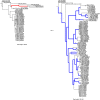
Flaviviruses display diverse epidemiological and ecological features. Tick-borne and mosquito-borne flaviviruses (TBFV and MBFV, respectively) are important human pathogens that alternate replication in invertebrate vectors and vertebrate hosts. The Flavivirus genus also includes insect-specific viruses (ISFVs) and viruses with unknown invertebrate hosts. It is generally accepted that viruses that alternate between taxonomically different hosts evolve slowly and that the evolution of MBFVs and TBFVs is dominated by strong constraints, with limited episodes of positive selection. We exploited the availability of flavivirus genomes to test these hypotheses and to compare their rates and patterns of evolution. We estimated the substitution rates of CFAV and CxFV (two ISFVs) and, by taking into account the time-frame of measurement, compared them with those of other flaviviruses. Results indicated that CFAV and CxFV display relatively different substitution rates. However, these data, together with estimates for single-host members of the Flaviviridae family, indicated that MBFVs do not display relatively slower evolution. Conversely, TBFVs displayed some of lowest substitution rates among flaviviruses. Analysis of selective patterns over longer evolutionary time-frames confirmed that MBFVs evolve under strong purifying selection. Interestingly, TBFVs and ISFVs did not show extremely different levels of constraint, although TBFVs alternate among hosts, whereas ISFVs do not. Additional results showed that episodic positive selection drove the evolution of MBFVs, despite their high constraint. Positive selection was also detected on two branches of the TBFVs phylogeny that define the seabird clade. Thus, positive selection was much more common during the evolution of arthropod-borne flaviviruses than previously thought. Overall, our data indicate that flavivirus evolutionary patterns are complex and most likely determined by multiple factors, not limited to the alternation between taxonomically divergent hosts. The frequency of both positive and purifying selection, especially in MBFVs, suggests that a minority of sites in the viral polyprotein experience weak constraint and can evolve to generate new viral phenotypes and possibly promote adaptation to new hosts.
Keywords: Dating analysis; Episodic positive selection; Flavivirus evolution; Hosts alternation; Mosquito-borne flavivirus; Tick-borne flavivirus.
© The Author(s) 2021. Published by Oxford University Press.
Figures


References
-
- Abushouk A. I., Negida A., Ahmed H. (2016) ‘An Updated Review of Zika Virus’, Journal of Clinical Virology : The Official Publication of the Pan American Society for Clinical Virology, 84: 53–8. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

