A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
- PMID: 33977052
- PMCID: PMC8097335
- DOI: 10.1002/advs.202003630
A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Abstract
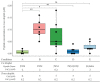
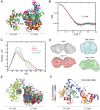
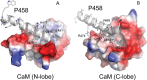
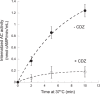
The molecular mechanisms and forces involved in the translocation of bacterial toxins into host cells are still a matter of intense research. The adenylate cyclase (CyaA) toxin from Bordetella pertussis displays a unique intoxication pathway in which its catalytic domain is directly translocated across target cell membranes. The CyaA translocation region contains a segment, P454 (residues 454-484), which exhibits membrane-active properties related to antimicrobial peptides. Herein, the results show that this peptide is able to translocate across membranes and to interact with calmodulin (CaM). Structural and biophysical analyses reveal the key residues of P454 involved in membrane destabilization and calmodulin binding. Mutational analysis demonstrates that these residues play a crucial role in CyaA translocation into target cells. In addition, calmidazolium, a calmodulin inhibitor, efficiently blocks CyaA internalization. It is proposed that after CyaA binding to target cells, the P454 segment destabilizes the plasma membrane, translocates across the lipid bilayer and binds calmodulin. Trapping of CyaA by the CaM:P454 interaction in the cytosol may assist the entry of the N-terminal catalytic domain by converting the stochastic motion of the polypeptide chain through the membrane into an efficient vectorial chain translocation into host cells.
Keywords: Bordetella pertussis; CyaA toxin; adenylate cyclase; calmodulin; membrane translocation; protein membrane interaction; protein–protein interactions.
© 2021 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Coote J. G., FEMS Microbiol. Rev. 1992, 8, 137. - PubMed
-
- Chenal A., Sotomayor Perez A. C., Ladant D., in The Comprehensive Sourcebook of Bacterial Protein Toxins, 4th ed. (Eds: Alouf J., Ladant D., Popoff M. R.), Elsevier, Amsterdam, Netherlands: 2015.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
