Old yeasts, young beer-The industrial relevance of yeast chronological life span
- PMID: 33978982
- PMCID: PMC8252602
- DOI: 10.1002/yea.3650
Old yeasts, young beer-The industrial relevance of yeast chronological life span
Abstract
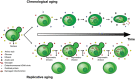
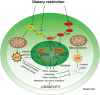
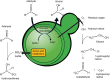
Much like other living organisms, yeast cells have a limited life span, in terms of both the maximal length of time a cell can stay alive (chronological life span) and the maximal number of cell divisions it can undergo (replicative life span). Over the past years, intensive research revealed that the life span of yeast depends on both the genetic background of the cells and environmental factors. Specifically, the presence of stress factors, reactive oxygen species, and the availability of nutrients profoundly impact life span, and signaling cascades involved in the response to these factors, including the target of rapamycin (TOR) and cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA) pathways, play a central role. Interestingly, yeast life span also has direct implications for its use in industrial processes. In beer brewing, for example, the inoculation of finished beer with live yeast cells, a process called "bottle conditioning" helps improve the product's shelf life by clearing undesirable carbonyl compounds such as furfural and 2-methylpropanal that cause staling. However, this effect depends on the reductive metabolism of living cells and is thus inherently limited by the cells' chronological life span. Here, we review the mechanisms underlying chronological life span in yeast. We also discuss how this insight connects to industrial observations and ultimately opens new routes towards superior industrial yeasts that can help improve a product's shelf life and thus contribute to a more sustainable industry.
Keywords: PKA pathway; TORC1/Sch9; bottle conditioning; chronological life span; flavor stability; yeast.
© 2021 The Authors. Yeast published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors are presently engaged in research aimed at generating yeasts with a superior chronological life span for industrial use, where the laboratories and project are in part financially funded by Duvel Moortgat, which may eventually lead to the development of products that may be patent protected and licensed.
Figures
References
-
- Ahrens, H. , Schrpfer, J. , Stumpf, L. , Pahl, R. , Brauer, J. , & Schildbach, S. (2018a). Enhancing flavour stability in beer using biological scavengers part 1: Methodology and preliminary trials. BrewingScience, 71(1–2), 12–17. 10.23763/BrSc18-04schildbach - DOI
-
- Ahrens, H. , Schrpfer, J. , Stumpf, L. , Pahl, R. , Brauer, J. , & Schildbach, S. (2018b). Enhancing flavour stability in beer using biological scavengers part 2: Screening of yeasts. BrewingScience, 71(3–4), 24–30. 10.23763/BrSc18-04schildbach - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
