Quantum Chemistry Calculations for Metabolomics
- PMID: 33979149
- PMCID: PMC8161423
- DOI: 10.1021/acs.chemrev.0c00901
Quantum Chemistry Calculations for Metabolomics
Abstract
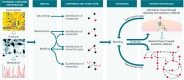
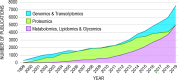
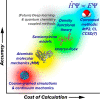
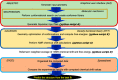
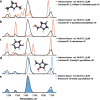
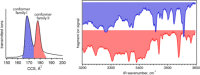
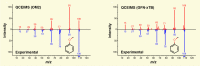
A primary goal of metabolomics studies is to fully characterize the small-molecule composition of complex biological and environmental samples. However, despite advances in analytical technologies over the past two decades, the majority of small molecules in complex samples are not readily identifiable due to the immense structural and chemical diversity present within the metabolome. Current gold-standard identification methods rely on reference libraries built using authentic chemical materials ("standards"), which are not available for most molecules. Computational quantum chemistry methods, which can be used to calculate chemical properties that are then measured by analytical platforms, offer an alternative route for building reference libraries, i.e., in silico libraries for "standards-free" identification. In this review, we cover the major roadblocks currently facing metabolomics and discuss applications where quantum chemistry calculations offer a solution. Several successful examples for nuclear magnetic resonance spectroscopy, ion mobility spectrometry, infrared spectroscopy, and mass spectrometry methods are reviewed. Finally, we consider current best practices, sources of error, and provide an outlook for quantum chemistry calculations in metabolomics studies. We expect this review will inspire researchers in the field of small-molecule identification to accelerate adoption of in silico methods for generation of reference libraries and to add quantum chemistry calculations as another tool at their disposal to characterize complex samples.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


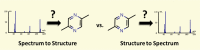


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

