Generation of a DAT-P2A-Flpo mouse line for intersectional genetic targeting of dopamine neuron subpopulations
- PMID: 33979604
- PMCID: PMC8240967
- DOI: 10.1016/j.celrep.2021.109123
Generation of a DAT-P2A-Flpo mouse line for intersectional genetic targeting of dopamine neuron subpopulations
Abstract
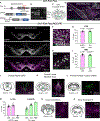
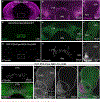
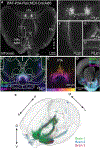
Dopaminergic projections exert widespread influence over multiple brain regions and modulate various behaviors including movement, reward learning, and motivation. It is increasingly appreciated that dopamine neurons are heterogeneous in their gene expression, circuitry, physiology, and function. Current approaches to target dopamine neurons are largely based on single gene drivers, which either label all dopamine neurons or mark a subset but concurrently label non-dopaminergic neurons. Here, we establish a mouse line with Flpo recombinase expressed from the endogenous Slc6a3 (dopamine active transporter [DAT]) locus. DAT-P2A-Flpo mice can be used together with Cre-expressing mouse lines to efficiently and selectively label dopaminergic subpopulations using Cre/Flp-dependent intersectional strategies. We demonstrate the utility of this approach by generating DAT-P2A-Flpo;NEX-Cre mice that specifically label Neurod6-expressing dopamine neurons, which project to the nucleus accumbens medial shell. DAT-P2A-Flpo mice add to a growing toolbox of genetic resources that will help parse the diverse functions mediated by dopaminergic circuits.
Keywords: DAT; DAT-dopamine neurons; Flp recombinase; Neurod6; TrailMap; intersectional genetics; nucleus accumbens; substantia nigra pars compacta; ventral tegmental area.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no financial or non-financial competing interests.
Figures


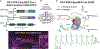
References
-
- Agarwal A, Dibaj P, Kassmann CM, Goebbels S, Nave KA, and Schwab MH (2012). In vivo imaging and noninvasive ablation of pyramidal neurons in adult NEX-CreERT2 mice. Cereb. Cortex 22, 1473–1486. - PubMed
-
- Anderzhanova EA, Bächli H, Buneeva OA, Narkevich VB, Medvedev AE, Thoeringer CK, Wotjak CT, and Kudrin VS (2013). Strain differences in profiles of dopaminergic neurotransmission in the prefrontal cortex of the BALB/C vs. C57Bl/6 mice: consequences of stress and afobazole. Eur. J. Pharmacol. 708, 95–104. - PubMed
-
- Awatramani R, Soriano P, Rodriguez C, Mai JJ, and Dymecki SM (2003). Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat. Genet. 35, 70–75. - PubMed
-
- Bäckman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, Westphal H, and Tomac AC (2006). Characterization of a mouse strain expressing Cre recombinase from the 3′ untranslated region of the dopamine transporter locus. Genesis 44, 383–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

