Chromatin accessibility governs the differential response of cancer and T cells to arginine starvation
- PMID: 33979616
- PMCID: PMC8131582
- DOI: 10.1016/j.celrep.2021.109101
Chromatin accessibility governs the differential response of cancer and T cells to arginine starvation
Abstract
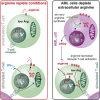
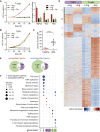
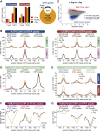
Depleting the microenvironment of important nutrients such as arginine is a key strategy for immune evasion by cancer cells. Many tumors overexpress arginase, but it is unclear how these cancers, but not T cells, tolerate arginine depletion. In this study, we show that tumor cells synthesize arginine from citrulline by upregulating argininosuccinate synthetase 1 (ASS1). Under arginine starvation, ASS1 transcription is induced by ATF4 and CEBPβ binding to an enhancer within ASS1. T cells cannot induce ASS1, despite the presence of active ATF4 and CEBPβ, as the gene is repressed. Arginine starvation drives global chromatin compaction and repressive histone methylation, which disrupts ATF4/CEBPβ binding and target gene transcription. We find that T cell activation is impaired in arginine-depleted conditions, with significant metabolic perturbation linked to incomplete chromatin remodeling and misregulation of key genes. Our results highlight a T cell behavior mediated by nutritional stress, exploited by cancer cells to enable pathological immune evasion.
Keywords: ASS1; ATF4; H3K27me3; T cell chromatin; arginine; cancer metabolism; immunometabolism; immunosuppression; metabolic regulation; nutritional stress.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.A.M. and P.V. are founder shareholders of OxStem Oncology (OSO), a subsidiary company of OxStem Ltd. M. Salio consults for Nucleome Therapeutics Ltd. The remaining authors declare no competing interests.
Figures





References
-
- Ameri K., Harris A.L. Activating transcription factor 4. Int. J. Biochem. Cell Biol. 2008;40:14–21. - PubMed
-
- Bateman L.A., Ku W.M., Heslin M.J., Contreras C.M., Skibola C.F., Nomura D.K. Argininosuccinate synthase 1 is a metabolic regulator of colorectal cancer pathogenicity. ACS Chem. Biol. 2017;12:905–911. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

