RIPK1 activation mediates neuroinflammation and disease progression in multiple sclerosis
- PMID: 33979622
- PMCID: PMC8917516
- DOI: 10.1016/j.celrep.2021.109112
RIPK1 activation mediates neuroinflammation and disease progression in multiple sclerosis
Abstract
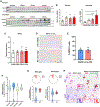
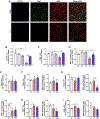
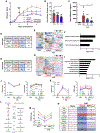
Receptor interacting protein kinase 1 (RIPK1) mediates cell death and inflammatory signaling and is increased in multiple sclerosis (MS) brain samples. Here, we investigate the role of glial RIPK1 kinase activity in mediating MS pathogenesis. We demonstrate RIPK1 levels correlate with MS disease progression. We find microglia are susceptible to RIPK1-mediated cell death and identify an inflammatory gene signature that may contribute to the neuroinflammatory milieu in MS patients. We uncover a distinct role for RIPK1 in astrocytes in regulating inflammatory signaling in the absence of cell death and confirm RIPK1-kinase-dependent regulation in human glia. Using a murine MS model, we show RIPK1 inhibition attenuates disease progression and suppresses deleterious signaling in astrocytes and microglia. Our results suggest RIPK1 kinase activation in microglia and astrocytes induces a detrimental neuroinflammatory program that contributes to the neurodegenerative environment in progressive MS.
Keywords: RIPK1; astrocyte; cell death; inflammation; microglia; multiple sclerosis; necroptosis.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.Z., F.P., L.W., C.Z., A.M., Y.R., M.L., R.G., L.G., P.L., T.X., T.H., and D.O. were employees of Sanofi at the time this research was conducted. The authors declare no other conflicts of interest.
Figures



References
-
- Baecher-Allan C, Kaskow BJ, and Weiner HL (2018). Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 97, 742–768. - PubMed
-
- Berard JL, Wolak K, Fournier S, and David S (2010). Characterization of relapsing-remitting and chronic forms of experimental autoimmune encephalomyelitis in C57BL/6 mice. Glia 58, 434–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

