Echinacea purpurea (L.) Moench treatment of monocytes promotes tonic interferon signaling, increased innate immunity gene expression and DNA repeat hypermethylated silencing of endogenous retroviral sequences
- PMID: 33980308
- PMCID: PMC8114977
- DOI: 10.1186/s12906-021-03310-5
Echinacea purpurea (L.) Moench treatment of monocytes promotes tonic interferon signaling, increased innate immunity gene expression and DNA repeat hypermethylated silencing of endogenous retroviral sequences
Abstract
Background: Herbal remedies of Echinacea purpurea tinctures are widely used today to reduce common cold respiratory tract infections.
Methods: Transcriptome, epigenome and kinome profiling allowed a systems biology level characterisation of genomewide immunomodulatory effects of a standardized Echinacea purpurea (L.) Moench extract in THP1 monocytes.
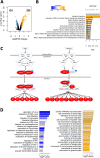
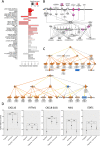
Results: Gene expression and DNA methylation analysis revealed that Echinaforce® treatment triggers antiviral innate immunity pathways, involving tonic IFN signaling, activation of pattern recognition receptors, chemotaxis and immunometabolism. Furthermore, phosphopeptide based kinome activity profiling and pharmacological inhibitor experiments with filgotinib confirm a key role for Janus Kinase (JAK)-1 dependent gene expression changes in innate immune signaling. Finally, Echinaforce® treatment induces DNA hypermethylation at intergenic CpG, long/short interspersed nuclear DNA repeat elements (LINE, SINE) or long termininal DNA repeats (LTR). This changes transcription of flanking endogenous retroviral sequences (HERVs), involved in an evolutionary conserved (epi) genomic protective response against viral infections.
Conclusions: Altogether, our results suggest that Echinaforce® phytochemicals strengthen antiviral innate immunity through tonic IFN regulation of pattern recognition and chemokine gene expression and DNA repeat hypermethylated silencing of HERVs in monocytes. These results suggest that immunomodulation by Echinaforce® treatment holds promise to reduce symptoms and duration of infection episodes of common cold corona viruses (CoV), Severe Acute Respiratory Syndrome (SARS)-CoV, and new occurring strains such as SARS-CoV-2, with strongly impaired interferon (IFN) response and weak innate antiviral defense.
Conflict of interest statement
Author Andreas Suter was employed by the company A. Vogel Bioforce AG, Roggwil, Switzerland. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Echinacea Purpurea For the Long-Term Prevention of Viral Respiratory Tract Infections During Covid-19 Pandemic: A Randomized, Open, Controlled, Exploratory Clinical Study.Front Pharmacol. 2022 Apr 26;13:856410. doi: 10.3389/fphar.2022.856410. eCollection 2022. Front Pharmacol. 2022. PMID: 35559249 Free PMC article.
-
Efficacy and safety of Echinaforce® in respiratory tract infections.Wien Med Wochenschr. 2013 Feb;163(3-4):102-5. doi: 10.1007/s10354-012-0166-0. Epub 2012 Dec 20. Wien Med Wochenschr. 2013. PMID: 23263637 Free PMC article. Review.
-
Echinacea purpurea: A Proprietary Extract of Echinacea purpurea Is Shown to be Safe and Effective in the Prevention of the Common Cold.Holist Nurs Pract. 2016 Jan-Feb;30(1):54-7. doi: 10.1097/HNP.0000000000000130. Holist Nurs Pract. 2016. PMID: 26633727 Review.
-
Changes in immunomodulatory properties of Echinacea spp. root infusions and tinctures stored at 4 degrees C for four days.Clin Chim Acta. 2005 May;355(1-2):67-82. doi: 10.1016/j.cccn.2004.12.013. Clin Chim Acta. 2005. PMID: 15820480
-
Bactericidal and anti-inflammatory properties of a standardized Echinacea extract (Echinaforce): dual actions against respiratory bacteria.Phytomedicine. 2010 Jul;17(8-9):563-8. doi: 10.1016/j.phymed.2009.10.022. Epub 2009 Dec 29. Phytomedicine. 2010. PMID: 20036523
Cited by
-
Echinacea Purpurea For the Long-Term Prevention of Viral Respiratory Tract Infections During Covid-19 Pandemic: A Randomized, Open, Controlled, Exploratory Clinical Study.Front Pharmacol. 2022 Apr 26;13:856410. doi: 10.3389/fphar.2022.856410. eCollection 2022. Front Pharmacol. 2022. PMID: 35559249 Free PMC article.
-
Echinacea Reduces Antibiotics by Preventing Respiratory Infections: A Meta-Analysis (ERA-PRIMA).Antibiotics (Basel). 2024 Apr 16;13(4):364. doi: 10.3390/antibiotics13040364. Antibiotics (Basel). 2024. PMID: 38667040 Free PMC article. Review.
-
Echinacea as a Potential Force against Coronavirus Infections? A Mini-Review of Randomized Controlled Trials in Adults and Children.Microorganisms. 2022 Jan 19;10(2):211. doi: 10.3390/microorganisms10020211. Microorganisms. 2022. PMID: 35208665 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

