Selective Requirement of MYB for Oncogenic Hyperactivation of a Translocated Enhancer in Leukemia
- PMID: 33980539
- PMCID: PMC8563373
- DOI: 10.1158/2159-8290.CD-20-1793
Selective Requirement of MYB for Oncogenic Hyperactivation of a Translocated Enhancer in Leukemia
Abstract
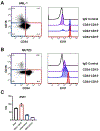
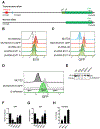
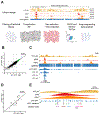
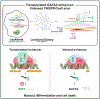
In acute myeloid leukemia (AML) with inv(3)(q21;q26) or t(3;3)(q21;q26), a translocated GATA2 enhancer drives oncogenic expression of EVI1. We generated an EVI1-GFP AML model and applied an unbiased CRISPR/Cas9 enhancer scan to uncover sequence motifs essential for EVI1 transcription. Using this approach, we pinpointed a single regulatory element in the translocated GATA2 enhancer that is critically required for aberrant EVI1 expression. This element contained a DNA-binding motif for the transcription factor MYB, which specifically occupied this site at the translocated allele and was dispensable for GATA2 expression. MYB knockout as well as peptidomimetic blockade of CBP/p300-dependent MYB functions resulted in downregulation of EVI1 but not of GATA2. Targeting MYB or mutating its DNA-binding motif within the GATA2 enhancer resulted in myeloid differentiation and cell death, suggesting that interference with MYB-driven EVI1 transcription provides a potential entry point for therapy of inv(3)/t(3;3) AMLs. SIGNIFICANCE: We show a novel paradigm in which chromosomal aberrations reveal critical regulatory elements that are nonfunctional at their endogenous locus. This knowledge provides a rationale to develop new compounds to selectively interfere with oncogenic enhancer activity.This article is highlighted in the In This Issue feature, p. 2659.
©2021 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest disclosure
A patent application related to MYBMIM has been submitted by A.K. to the U.S. Patent and Trademark Office entitled “Agents and methods for treating CREB binding protein-dependent cancers” (application PCT/US2017/059579). A.K. received personal fees from Novartis and from Rgenta during the conduct of the study.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

