Innate Antiviral Cytokine Response to Swine Influenza Virus by Swine Respiratory Epithelial Cells
- PMID: 33980596
- PMCID: PMC8274599
- DOI: 10.1128/JVI.00692-21
Innate Antiviral Cytokine Response to Swine Influenza Virus by Swine Respiratory Epithelial Cells
Abstract
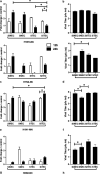
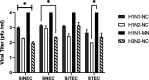
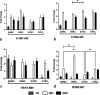
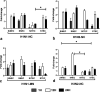
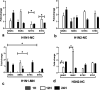
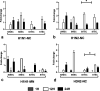
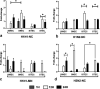
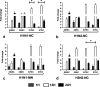
Swine influenza virus (SIV) can cause respiratory illness in swine. Swine contribute to influenza virus reassortment, as avian, human, and/or swine influenza viruses can infect swine and reassort, and new viruses can emerge. Thus, it is important to determine the host antiviral responses that affect SIV replication. In this study, we examined the innate antiviral cytokine response to SIV by swine respiratory epithelial cells, focusing on the expression of interferon (IFN) and interferon-stimulated genes (ISGs). Both primary and transformed swine nasal and tracheal respiratory epithelial cells were examined following infection with field isolates. The results show that IFN and ISG expression is maximal at 12 h postinfection (hpi) and is dependent on cell type and virus genotype. IMPORTANCE Swine are considered intermediate hosts that have facilitated influenza virus reassortment events that have given rise pandemics or genetically related viruses have become established in swine. In this study, we examine the innate antiviral response to swine influenza virus in primary and immortalized swine nasal and tracheal epithelial cells, and show virus strain- and host cell type-dependent differential expression of key interferons and interferon-stimulated genes.
Keywords: interferon-stimulated gene; interferons; swine influenza virus; swine nasal cells; swine tracheal cells.
Figures
References
-
- Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed). 2013. Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

