Distinct mutational backgrounds and clonal architectures implicated prognostic discrepancies in small-cell carcinomas of the esophagus and lung
- PMID: 33980813
- PMCID: PMC8115141
- DOI: 10.1038/s41419-021-03754-0
Distinct mutational backgrounds and clonal architectures implicated prognostic discrepancies in small-cell carcinomas of the esophagus and lung
Abstract
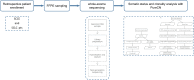
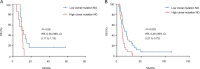
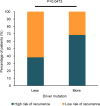
Small-cell carcinoma of the esophagus (SCCE) is a rare and aggressive cancer. Although several consistent genomic changes were observed previously between SCCE and small-cell lung cancer (SCLC), detailed mutational landscapes revealing discrepancies in genetic underpinnings of tumorigenesis between these two cancers are scarce, and little attention has been paid to answer whether these genetic alterations were related to the prognosis. Herein by performing whole-exome sequencing of 48 SCCE and 64 SCLC tumor samples, respectively we have shown that the number of driver mutations in SCCE was significantly lower than in SCLC (p = 0.0042). In SCCE, 46% of recurrent driver mutations were clonal, which occurred at an early stage during tumorigenesis, while 16 driver mutations were found clonal in SCLC. NOTCH1/3, PIK3CA, and ATM were specifically clonal in SCCE, while TP53 was clonal in SCLC. The total number of clonal mutations differed between two cancers and presented lower in SCCE compared to SCLC (p = 0.0036). Moreover, overall survival (OS) was shorter in patients with higher numbers of clonal mutations for both cancers. In summary, SCCE showed distinct mutational background and clonal architecture compared with SCLC. Organ-specific clonal events revealed different molecular mechanisms underlying tumorigenesis, tumor development, patients' prognosis, and possible variations in therapeutic outcomes to candidate treatments.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

