PD-1 suppresses TCR-CD8 cooperativity during T-cell antigen recognition
- PMID: 33980853
- PMCID: PMC8115078
- DOI: 10.1038/s41467-021-22965-9
PD-1 suppresses TCR-CD8 cooperativity during T-cell antigen recognition
Abstract
Despite the clinical success of blocking its interactions, how PD-1 inhibits T-cell activation is incompletely understood, as exemplified by its potency far exceeding what might be predicted from its affinity for PD-1 ligand-1 (PD-L1). This may be partially attributed to PD-1's targeting the proximal signaling of the T-cell receptor (TCR) and co-stimulatory receptor CD28 via activating Src homology region 2 domain-containing phosphatases (SHPs). Here, we report PD-1 signaling regulates the initial TCR antigen recognition manifested in a smaller spreading area, fewer molecular bonds formed, and shorter bond lifetime of T cell interaction with peptide-major histocompatibility complex (pMHC) in the presence than absence of PD-L1 in a manner dependent on SHPs and Leukocyte C-terminal Src kinase. Our results identify a PD-1 inhibitory mechanism that disrupts the cooperative TCR-pMHC-CD8 trimolecular interaction, which prevents CD8 from augmenting antigen recognition, explaining PD-1's potent inhibitory function and its value as a target for clinical intervention.
Conflict of interest statement
The authors declare no competing interests.
Figures
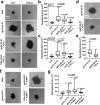
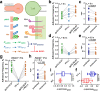

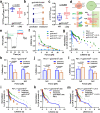
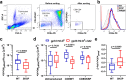
References
-
- Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc. Natl Acad. Sci. USA. 2001;98:13866–13871. doi: 10.1073/pnas.231486598. - DOI - PMC - PubMed
-
- Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol. 2004;173:945–954. doi: 10.4049/jimmunol.173.2.945. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

