Evaluation of the association of heterozygous germline variants in NTHL1 with breast cancer predisposition: an international multi-center study of 47,180 subjects
- PMID: 33980861
- PMCID: PMC8115524
- DOI: 10.1038/s41523-021-00255-3
Evaluation of the association of heterozygous germline variants in NTHL1 with breast cancer predisposition: an international multi-center study of 47,180 subjects
Abstract
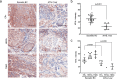
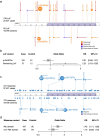
Bi-allelic loss-of-function (LoF) variants in the base excision repair (BER) gene NTHL1 cause a high-risk hereditary multi-tumor syndrome that includes breast cancer, but the contribution of heterozygous variants to hereditary breast cancer is unknown. An analysis of 4985 women with breast cancer, enriched for familial features, and 4786 cancer-free women revealed significant enrichment for NTHL1 LoF variants. Immunohistochemistry confirmed reduced NTHL1 expression in tumors from heterozygous carriers but the NTHL1 bi-allelic loss characteristic mutational signature (SBS 30) was not present. The analysis was extended to 27,421 breast cancer cases and 19,759 controls from 10 international studies revealing 138 cases and 93 controls with a heterozygous LoF variant (OR 1.06, 95% CI: 0.82-1.39) and 316 cases and 179 controls with a missense variant (OR 1.31, 95% CI: 1.09-1.57). Missense variants selected for deleterious features by a number of in silico bioinformatic prediction tools or located within the endonuclease III functional domain showed a stronger association with breast cancer. Somatic sequencing of breast cancers from carriers indicated that the risk associated with NTHL1 appears to operate through haploinsufficiency, consistent with other described low-penetrance breast cancer genes. Data from this very large international multicenter study suggests that heterozygous pathogenic germline coding variants in NTHL1 may be associated with low- to moderate- increased risk of breast cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Adenomas from individuals with pathogenic biallelic variants in the MUTYH and NTHL1 genes demonstrate base excision repair tumour mutational signature profiles similar to colorectal cancers, expanding potential diagnostic and variant classification applications.medRxiv [Preprint]. 2024 Aug 9:2024.08.08.24311713. doi: 10.1101/2024.08.08.24311713. medRxiv. 2024. Update in: Transl Oncol. 2025 Feb;52:102266. doi: 10.1016/j.tranon.2024.102266. PMID: 39148833 Free PMC article. Updated. Preprint.
-
Prevalence and Characterization of Biallelic and Monoallelic NTHL1 and MSH3 Variant Carriers From a Pan-Cancer Patient Population.JCO Precis Oncol. 2021 Feb 26;5:PO.20.00443. doi: 10.1200/PO.20.00443. eCollection 2021. JCO Precis Oncol. 2021. PMID: 34250384 Free PMC article.
-
Adenomas from individuals with pathogenic biallelic variants in the MUTYH and NTHL1 genes demonstrate base excision repair tumour mutational signature profiles similar to colorectal cancers, expanding potential diagnostic and variant classification applications.Transl Oncol. 2025 Feb;52:102266. doi: 10.1016/j.tranon.2024.102266. Epub 2025 Jan 9. Transl Oncol. 2025. PMID: 39793275 Free PMC article.
-
NTHL1 Tumor Syndrome.2020 Apr 2 [updated 2025 Mar 20]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2020 Apr 2 [updated 2025 Mar 20]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 32239880 Free Books & Documents. Review.
-
POLE, POLD1, and NTHL1: the last but not the least hereditary cancer-predisposing genes.Oncogene. 2021 Oct;40(40):5893-5901. doi: 10.1038/s41388-021-01984-2. Epub 2021 Aug 6. Oncogene. 2021. PMID: 34363023 Review.
Cited by
-
How many is too many? Polyposis syndromes and what to do next.Curr Opin Gastroenterol. 2022 Jan 1;38(1):39-47. doi: 10.1097/MOG.0000000000000796. Curr Opin Gastroenterol. 2022. PMID: 34839308 Free PMC article. Review.
-
Cohort Profile: Lifepool.Int J Epidemiol. 2025 Apr 12;54(3):dyaf044. doi: 10.1093/ije/dyaf044. Int J Epidemiol. 2025. PMID: 40246327 Free PMC article. No abstract available.
-
Assessment of candidate high-grade serous ovarian carcinoma predisposition genes through integrated germline and tumour sequencing.NPJ Genom Med. 2025 Jan 10;10(1):1. doi: 10.1038/s41525-024-00447-3. NPJ Genom Med. 2025. PMID: 39794353 Free PMC article.
-
Genetic analyses of DNA repair pathway associated genes implicate new candidate cancer predisposing genes in ancestrally defined ovarian cancer cases.Front Oncol. 2023 Mar 8;13:1111191. doi: 10.3389/fonc.2023.1111191. eCollection 2023. Front Oncol. 2023. PMID: 36969007 Free PMC article.
-
Base-Excision Repair Mutational Signature in Two Sebaceous Carcinomas of the Eyelid.Genes (Basel). 2023 Nov 8;14(11):2055. doi: 10.3390/genes14112055. Genes (Basel). 2023. PMID: 38002998 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

