Application of multiple omics and network projection analyses to drug repositioning for pathogenic mosquito-borne viruses
- PMID: 33980888
- PMCID: PMC8115341
- DOI: 10.1038/s41598-021-89171-x
Application of multiple omics and network projection analyses to drug repositioning for pathogenic mosquito-borne viruses
Abstract
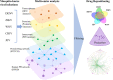
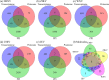
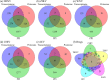
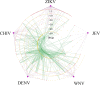
Pathogenic mosquito-borne viruses are a serious public health issue in tropical and subtropical regions and are increasingly becoming a problem in other climate zones. Drug repositioning is a rapid, pharmaco-economic approach that can be used to identify compounds that target these neglected tropical diseases. We have applied a computational drug repositioning method to five mosquito-borne viral infections: dengue virus (DENV), zika virus (ZIKV), West Nile virus (WNV), Japanese encephalitis virus (JEV) and Chikungunya virus (CHIV). We identified signature molecules and pathways for each virus infection based on omics analyses, and determined 77 drug candidates and 146 proteins for those diseases by using a filtering method. Based on the omics analyses, we analyzed the relationship among drugs, target proteins and the five viruses by projecting the signature molecules onto a human protein-protein interaction network. We have classified the drug candidates according to the degree of target proteins in the protein-protein interaction network for the five infectious diseases.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Neutralizing activities of human immunoglobulin derived from donors in Japan against mosquito-borne flaviviruses, Japanese encephalitis virus, West Nile virus, and dengue virus.Biologics. 2016 Jul 7;10:99-102. doi: 10.2147/BTT.S105650. eCollection 2016. Biologics. 2016. PMID: 27462140 Free PMC article.
-
Drug repositioning for dengue haemorrhagic fever by integrating multiple omics analyses.Sci Rep. 2019 Jan 24;9(1):523. doi: 10.1038/s41598-018-36636-1. Sci Rep. 2019. PMID: 30679503 Free PMC article.
-
Surveillance of mosquito-borne infectious diseases in febrile travelers entering China via Shenzhen ports, China, 2013.Travel Med Infect Dis. 2016 Mar-Apr;14(2):123-30. doi: 10.1016/j.tmaid.2016.02.002. Epub 2016 Mar 4. Travel Med Infect Dis. 2016. PMID: 26960752
-
Antiviral Compounds for Blocking Arboviral Transmission in Mosquitoes.Viruses. 2021 Jan 14;13(1):108. doi: 10.3390/v13010108. Viruses. 2021. PMID: 33466915 Free PMC article. Review.
-
Role of autophagy during the replication and pathogenesis of common mosquito-borne flavi- and alphaviruses.Open Biol. 2019 Mar 29;9(3):190009. doi: 10.1098/rsob.190009. Open Biol. 2019. PMID: 30862253 Free PMC article. Review.
Cited by
-
Drug repositioning as a promising approach for the eradication of emerging and re-emerging viral agents.Mol Divers. 2025 Mar 18. doi: 10.1007/s11030-025-11131-8. Online ahead of print. Mol Divers. 2025. PMID: 40100484 Review.
-
Exploring Immune Cell Infiltration and Small Molecule Compounds for Ulcerative Colitis Treatment.Genes (Basel). 2024 Nov 29;15(12):1548. doi: 10.3390/genes15121548. Genes (Basel). 2024. PMID: 39766817 Free PMC article.
-
MixOmics Integration of Biological Datasets Identifies Highly Correlated Variables of COVID-19 Severity.Int J Mol Sci. 2025 May 15;26(10):4743. doi: 10.3390/ijms26104743. Int J Mol Sci. 2025. PMID: 40429886 Free PMC article.
-
Deciphering the similarities and disparities of molecular mechanisms behind respiratory epithelium response to HCoV-229E and SARS-CoV-2 and drug repurposing, a systems biology approach.Daru. 2024 Jun;32(1):215-235. doi: 10.1007/s40199-024-00507-0. Epub 2024 Apr 23. Daru. 2024. PMID: 38652363 Free PMC article.
-
Pharmacophore-Model-Based Drug Repurposing for the Identification of the Potential Inhibitors Targeting the Allosteric Site in Dengue Virus NS5 RNA-Dependent RNA Polymerase.Viruses. 2022 Aug 20;14(8):1827. doi: 10.3390/v14081827. Viruses. 2022. PMID: 36016449 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

