Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei
- PMID: 33980975
- PMCID: PMC8115239
- DOI: 10.1038/s41598-021-89334-w
Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei
Abstract
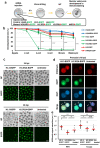
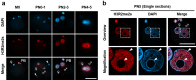
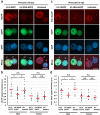
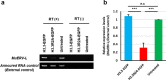
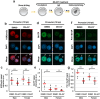
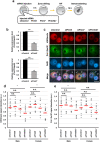
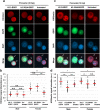
Paternal genome reprogramming, such as protamine-histone exchange and global DNA demethylation, is crucial for the development of fertilised embryos. Previously, our study showed that one of histone arginine methylation, asymmetrically dimethylated histone H3R17 (H3R17me2a), is necessary for epigenetic reprogramming in the mouse paternal genome. However, roles of histone arginine methylation in reprogramming after fertilisation are still poorly understood. Here, we report that H3R2me2s promotes global transcription at the 1-cell stage, referred to as minor zygotic genome activation (ZGA). The inhibition of H3R2me2s by expressing a histone H3.3 mutant H3.3R2A prevented embryonic development from the 2-cell to 4-cell stages and significantly reduced global RNA synthesis and RNA polymerase II (Pol II) activity. Consistent with this result, the expression levels of MuERV-L as minor ZGA transcripts were decreased by forced expression of H3.3R2A. Furthermore, treatment with an inhibitor and co-injection of siRNA to PRMT5 and PRMT7 also resulted in the attenuation of transcriptional activities with reduction of H3R2me2s in the pronuclei of zygotes. Interestingly, impairment of H3K4 methylation by expression of H3.3K4M resulted in a decrease of H3R2me2s in male pronuclei. Our findings suggest that H3R2me2s together with H3K4 methylation is involved in global transcription during minor ZGA in mice.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Exchanges of histone methylation and variants during mouse zygotic genome activation.Zygote. 2020 Feb;28(1):51-58. doi: 10.1017/S0967199419000649. Epub 2019 Nov 20. Zygote. 2020. PMID: 31746724
-
Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development.EMBO Rep. 2015 Jul;16(7):803-12. doi: 10.15252/embr.201439700. Epub 2015 Apr 29. EMBO Rep. 2015. PMID: 25925669 Free PMC article.
-
Use of Histone K-M Mutants for the Analysis of Transcriptional Regulation in Mouse Zygotes.Methods Mol Biol. 2017;1605:259-270. doi: 10.1007/978-1-4939-6988-3_18. Methods Mol Biol. 2017. PMID: 28456971
-
Epigenetic reprogramming of the zygote in mice and men: on your marks, get set, go!Reproduction. 2016 Dec;152(6):R211-R222. doi: 10.1530/REP-16-0376. Epub 2016 Sep 6. Reproduction. 2016. PMID: 27601712 Free PMC article. Review.
-
Understanding paternal genome demethylation through live-cell imaging and siRNA.Cell Mol Life Sci. 2011 May;68(10):1669-79. doi: 10.1007/s00018-010-0623-0. Epub 2011 Jan 15. Cell Mol Life Sci. 2011. PMID: 21234640 Free PMC article. Review.
Cited by
-
Dysregulation of arginine methylation in tumorigenesis.Front Mol Biosci. 2024 Jun 7;11:1420365. doi: 10.3389/fmolb.2024.1420365. eCollection 2024. Front Mol Biosci. 2024. PMID: 38911125 Free PMC article. Review.
-
Critical Roles of Protein Arginine Methylation in the Central Nervous System.Mol Neurobiol. 2023 Oct;60(10):6060-6091. doi: 10.1007/s12035-023-03465-x. Epub 2023 Jul 6. Mol Neurobiol. 2023. PMID: 37415067 Review.
-
Structure and Function of Protein Arginine Methyltransferase PRMT7.Life (Basel). 2021 Jul 30;11(8):768. doi: 10.3390/life11080768. Life (Basel). 2021. PMID: 34440512 Free PMC article. Review.
-
The Novel Role of Zfp296 in Mammalian Embryonic Genome Activation as an H3K9me3 Modulator.Int J Mol Sci. 2023 Jul 12;24(14):11377. doi: 10.3390/ijms241411377. Int J Mol Sci. 2023. PMID: 37511136 Free PMC article.
-
The PRMT5-LSD1 axis confers Slug dual transcriptional activities and promotes breast cancer progression.J Exp Clin Cancer Res. 2022 Jun 2;41(1):191. doi: 10.1186/s13046-022-02400-7. J Exp Clin Cancer Res. 2022. PMID: 35655230 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

