Cell surface GRP78 and Dermcidin cooperate to regulate breast cancer cell migration through Wnt signaling
- PMID: 33981001
- PMCID: PMC8197743
- DOI: 10.1038/s41388-021-01821-6
Cell surface GRP78 and Dermcidin cooperate to regulate breast cancer cell migration through Wnt signaling
Abstract
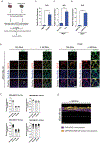
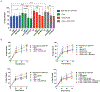
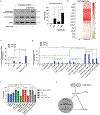
The heat shock protein GRP78 typically resides in the endoplasmic reticulum in normal tissues, but it has been shown to be expressed on the cell surface of several cancer cells, and some stem cells, where it can act as a signaling molecule by not-yet-fully defined mechanisms. Although cell surface GRP78 (sGRP78) has emerged as an attractive chemotherapeutic target, understanding how sGRP78 is functioning in cancer has been complicated by the fact that sGRP78 can function in a cell-context dependent manner, with a diverse array of reported binding partners, to regulate a variety of cellular responses. We had previously shown that sGRP78 was important in regulating pluripotent stem cell (PSC) functions, and hypothesized that embryonic-like mechanisms of GRP78 were critical to regulating aggressive breast cancer cell functions. Here, using proteomics we identify Dermcidin (DCD) as a novel sGRP78 binding partner common to both PSCs and breast cancer cells. We show that GRP78 and DCD cooperate to regulate stem cell and cancer cell migration that is dependent on the cell surface functions of these proteins. Finally, we identify Wnt/β-catenin signaling, a critical pathway in stem cell and cancer cell biology, as an important downstream intermediate in regulating this migration phenotype.
Conflict of interest statement
DISCLOSURE OF CONFLICTS OF INTEREST
The authors declare no competing interests.
Figures

References
-
- Chaffer CL, Weinberg RA: A perspective on cancer cell metastasis. Science 2011, 331:1559–1564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

