Pan-cancer association of HLA gene expression with cancer prognosis and immunotherapy efficacy
- PMID: 33981015
- PMCID: PMC8329209
- DOI: 10.1038/s41416-021-01400-2
Pan-cancer association of HLA gene expression with cancer prognosis and immunotherapy efficacy
Abstract
Background: The function of major histocompatibility complex (MHC) molecules is to bind peptide fragments derived from genomic mutations or pathogens and display them on the cell surface for recognition by cognate T cells to initiate an immune response.
Methods: In this study, we provide a comprehensive investigation of HLA gene expression in a pan-cancer manner involving 33 cancer types. We utilised gene expression data from several databases and immune checkpoint blockade-treated patient cohorts.
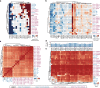
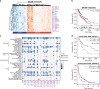
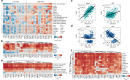
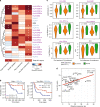
Results: We show that MHC expression varies strongly among cancer types and is associated with several genomic and immunological features. While immune cell infiltration was generally higher in tumours with higher HLA gene expression, CD4+ T cells showed significantly different correlations among cancer types, separating them into two clusters. Furthermore, we show that increased HLA gene expression is associated with prolonged survival in the majority of cancer types. Lastly, HLA gene expression is associated with patient response to immune checkpoint blockade, which is especially prominent for HLA class II expression in tumour biopsies taken during treatment.
Conclusion: We show that HLA gene expression is an important feature of tumour biology that has significant impact on patient prognosis.
© 2021. The Author(s), under exclusive licence to Cancer Research UK.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Albitar M, Johnson M, Do KA, Day A, Jilani I, Pierce S, et al. Levels of soluble HLA-I and beta2M in patients with acute myeloid leukemia and advanced myelodysplastic syndrome: association with clinical behavior and outcome of induction therapy. Leukemia. 2007;21:480–488. doi: 10.1038/sj.leu.2404506. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

