DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer
- PMID: 33981038
- PMCID: PMC8895686
- DOI: 10.1038/s41586-021-03539-7
DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer
Erratum in
-
Author Correction: DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer.Nature. 2021 Aug;596(7873):E13. doi: 10.1038/s41586-021-03820-9. Nature. 2021. PMID: 34341547 No abstract available.
Abstract
Ferroptosis, a form of regulated cell death that is induced by excessive lipid peroxidation, is a key tumour suppression mechanism1-4. Glutathione peroxidase 4 (GPX4)5,6 and ferroptosis suppressor protein 1 (FSP1)7,8 constitute two major ferroptosis defence systems. Here we show that treatment of cancer cells with GPX4 inhibitors results in acute depletion of N-carbamoyl-L-aspartate, a pyrimidine biosynthesis intermediate, with concomitant accumulation of uridine. Supplementation with dihydroorotate or orotate-the substrate and product of dihydroorotate dehydrogenase (DHODH)-attenuates or potentiates ferroptosis induced by inhibition of GPX4, respectively, and these effects are particularly pronounced in cancer cells with low expression of GPX4 (GPX4low). Inactivation of DHODH induces extensive mitochondrial lipid peroxidation and ferroptosis in GPX4low cancer cells, and synergizes with ferroptosis inducers to induce these effects in GPX4high cancer cells. Mechanistically, DHODH operates in parallel to mitochondrial GPX4 (but independently of cytosolic GPX4 or FSP1) to inhibit ferroptosis in the mitochondrial inner membrane by reducing ubiquinone to ubiquinol (a radical-trapping antioxidant with anti-ferroptosis activity). The DHODH inhibitor brequinar selectively suppresses GPX4low tumour growth by inducing ferroptosis, whereas combined treatment with brequinar and sulfasalazine, an FDA-approved drug with ferroptosis-inducing activity, synergistically induces ferroptosis and suppresses GPX4high tumour growth. Our results identify a DHODH-mediated ferroptosis defence mechanism in mitochondria and suggest a therapeutic strategy of targeting ferroptosis in cancer treatment.
Conflict of interest statement
Figures

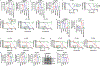


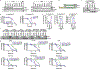





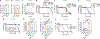
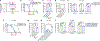

Comment in
-
A mitochondrial gatekeeper that helps cells escape death by ferroptosis.Nature. 2021 May;593(7860):514-515. doi: 10.1038/d41586-021-01203-8. Nature. 2021. PMID: 33981062 No abstract available.
-
DHODH inhibitors sensitize to ferroptosis by FSP1 inhibition.Nature. 2023 Jul;619(7968):E9-E18. doi: 10.1038/s41586-023-06269-0. Epub 2023 Jul 5. Nature. 2023. PMID: 37407687 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

