Mitochondrial TNAP controls thermogenesis by hydrolysis of phosphocreatine
- PMID: 33981039
- PMCID: PMC8287965
- DOI: 10.1038/s41586-021-03533-z
Mitochondrial TNAP controls thermogenesis by hydrolysis of phosphocreatine
Abstract
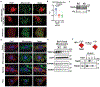
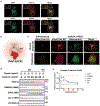
Adaptive thermogenesis has attracted much attention because of its ability to increase systemic energy expenditure and to counter obesity and diabetes1-3. Recent data have indicated that thermogenic fat cells use creatine to stimulate futile substrate cycling, dissipating chemical energy as heat4,5. This model was based on the super-stoichiometric relationship between the amount of creatine added to mitochondria and the quantity of oxygen consumed. Here we provide direct evidence for the molecular basis of this futile creatine cycling activity in mice. Thermogenic fat cells have robust phosphocreatine phosphatase activity, which is attributed to tissue-nonspecific alkaline phosphatase (TNAP). TNAP hydrolyses phosphocreatine to initiate a futile cycle of creatine dephosphorylation and phosphorylation. Unlike in other cells, TNAP in thermogenic fat cells is localized to the mitochondria, where futile creatine cycling occurs. TNAP expression is powerfully induced when mice are exposed to cold conditions, and its inhibition in isolated mitochondria leads to a loss of futile creatine cycling. In addition, genetic ablation of TNAP in adipocytes reduces whole-body energy expenditure and leads to rapid-onset obesity in mice, with no change in movement or feeding behaviour. These data illustrate the critical role of TNAP as a phosphocreatine phosphatase in the futile creatine cycle.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing financial interests.
Figures












Comment in
-
Unravelling the molecular basis of futile creatine cycling.Nat Rev Endocrinol. 2021 Jul;17(7):381. doi: 10.1038/s41574-021-00513-1. Nat Rev Endocrinol. 2021. PMID: 34021276 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

