Salmonella Typhimurium Triggers Extracellular Traps Release in Murine Macrophages
- PMID: 33981627
- PMCID: PMC8107695
- DOI: 10.3389/fcimb.2021.639768
Salmonella Typhimurium Triggers Extracellular Traps Release in Murine Macrophages
Abstract
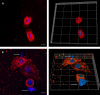
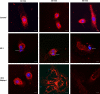
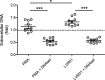
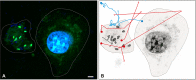
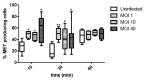
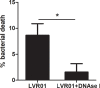
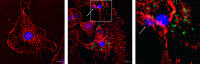
Salmonella comprises two species and more than 2500 serovars with marked differences in host specificity, and is responsible for a wide spectrum of diseases, ranging from localized gastroenteritis to severe life-threatening invasive disease. The initiation of the host inflammatory response, triggered by many Pathogen-Associated Molecular Patterns (PAMPs) that Salmonella possesses, recruits innate immune cells in order to restrain the infection at the local site. Neutrophils are known for killing bacteria through oxidative burst, amid other mechanisms. Amongst those mechanisms for controlling bacteria, the release of Extracellular Traps (ETs) represents a newly described pathway of programmed cell death known as ETosis. Particularly, Neutrophil Extracellular Traps (NETs) were first described in 2004 and since then, a number of reports have demonstrated their role as a novel defense mechanism against different pathogens. This released net-like material is composed of cellular DNA decorated with histones and cellular proteins. These structures have shown ability to trap, neutralize and kill different kinds of microorganisms, ranging from viruses and bacteria to fungi and parasites. Salmonella was one of the first microorganisms that were reported to be killed by NETs and several studies have confirmed the observation and deepened into its variants. Nevertheless, much less is known about their counterparts in other immune cells, e.g. Macrophage Extracellular Traps (METs) and Salmonella-induced MET release has never been reported so far. In this work, we observed the production of METs induced by Salmonella enterica serovar Typhimurium and recorded their effect on bacteria, showing for the first time that macrophages can also release extracellular DNA traps upon encounter with Salmonella Typhimurium. Additionally we show that METs effectively immobilize and reduce Salmonella survival in a few minutes, suggesting METs as a novel immune-mediated defense mechanism against Salmonella infection. Of note, this phenomenon was confirmed in primary macrophages, since MET release was also observed in bone marrow-derived macrophages infected with Salmonella. The evidence of this peculiar mechanism provides new incipient insights into macrophages´ role against Salmonella infection and can help to design new strategies for the clinical control of this transcendental pathogen.
Keywords: MET; Salmonella; defense mechanism; extracellular traps; macrophages.
Copyright © 2021 Mónaco, Canales-Huerta, Jara-Wilde, Härtel, Chabalgoity, Moreno and Scavone.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Azevedo E. P., Guimaraes-Costa A. B., Torezani G. S., Braga C. A., Palhano F. L., Kelly J. W., et al. . (2012). Amyloid Fibrils Trigger the Release of Neutrophil Extracellular Traps (Nets), Causing Fibril Fragmentation by NET-associated Elastase. J. Biol. Chem. 287 (44), 37206–37218. 10.1074/jbc.M112.369942 - DOI - PMC - PubMed
-
- Betancor L., Pereira M., Martinez A., Giossa G., Fookes M., Flores K., et al. . (2010). Prevalence of Salmonella Enterica in Poultry and Eggs in Uruguay During an Epidemic Due to Salmonella Enterica Serovar Enteritidis. J. Clin. Microbiol. 48 (7), 2413–2423. 10.1128/JCM.02137-09 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

