NMD inhibition by 5-azacytidine augments presentation of immunogenic frameshift-derived neoepitopes
- PMID: 33981976
- PMCID: PMC8082087
- DOI: 10.1016/j.isci.2021.102389
NMD inhibition by 5-azacytidine augments presentation of immunogenic frameshift-derived neoepitopes
Abstract
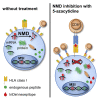
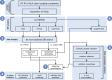
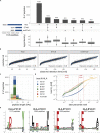
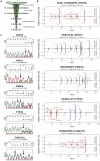
Frameshifted protein sequences elicit tumor-specific T cell-mediated immune responses in microsatellite-unstable (MSI) cancers if presented by HLA class I molecules. However, their expression and presentation are limited by nonsense-mediated RNA decay (NMD). We employed an unbiased immunopeptidomics workflow to analyze MSI HCT-116 cells and identified >10,000 HLA class I-presented peptides including five frameshift-derived InDel neoepitopes. Notably, pharmacological NMD inhibition with 5-azacytidine stabilizes frameshift-bearing transcripts and increases the HLA class I-mediated presentation of InDel neoepitopes. The frameshift mutation underlying one of the identified InDel neoepitopes is highly recurrent in MSI colorectal cancer cell lines and primary patient samples, and immunization with the corresponding neoepitope induces strong CD8+ T cell responses in an HLA-A∗02:01 transgenic mouse model. Our data show directly that pharmacological NMD inhibition augments HLA class I-mediated presentation of immunogenic frameshift-derived InDel neoepitopes thus highlighting the clinical potential of NMD inhibition in anti-cancer immunotherapy strategies.
Keywords: Biological Sciences; Cancer; Immunology.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Aliouat A., Hatin I., Bertin P., Francois P., Stierle V., Namy O., Salhi S., Jean-Jean O. Divergent effects of translation termination factor eRF3A and nonsense-mediated mRNA decay factor UPF1 on the expression of uORF carrying mRNAs and ribosome protein genes. RNA Biol. 2019;17:227–239. doi: 10.1080/15476286.2019.1674595. - DOI - PMC - PubMed
-
- Ballhausen A., Przybilla M.J., Jendrusch M., Haupt S., Pfaffendorf E., Seidler F., Witt J., Hernandez Sanchez A., Urban K., Draxlbauer M. The shared frameshift mutation landscape of microsatellite-unstable cancers suggests immunoediting during tumor evolution. Nat. Commun. 2020;11:4740. doi: 10.1038/s41467-020-18514-5. - DOI - PMC - PubMed
-
- Bassani-Sternberg M., Pletscher-Frankild S., Jensen L.J., Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell Proteomics. 2015;14:658–673. doi: 10.1074/mcp.M114.042812. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

